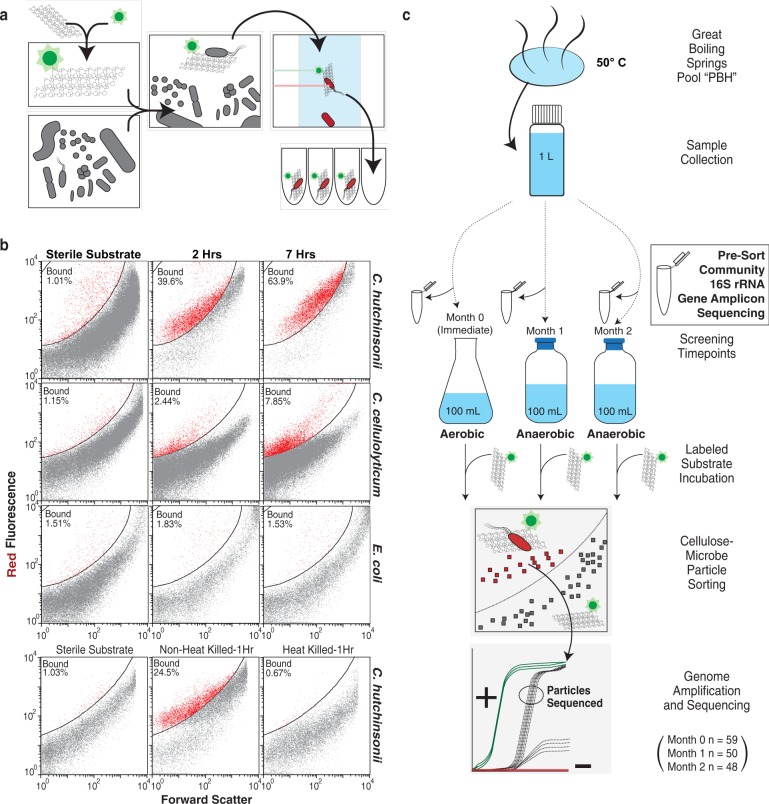
Fig. 1.
Function-driven single-cell genomics of cellulose-bound microbes. a Fluorescent substrate strategy used to identify microbes that colonize crystalline cellulose particles. The combination of a green fluorescent cellulose particle colonized by a red fluorescent DNA-stained microbe provides the signal to sort cellulose-adherent microbes from environmental samples. b Demonstration of flow cytometry gating strategy for identifying particles of labeled cellulose that become “bound” by microbes over time. Pure culture incubations of C. hutchinsonii (aerobic cellulose degrader), C. cellulolyticum (anaerobic cellulose degrader), and E. coli (noncellulose degrader) demonstrated specificity of binding to fluorescently labeled cellulose. Heat-killed C. hutchinsonii lost the ability to bind cellulose particles. c Experimental overview of function-driven sort and sequence data generated from Great Boiling Spring. Community composition of each bulk material (presort) was determined using 16S rRNA gene amplicon sequencing. Following incubation with labeled cellulose, cellulose-microbe particles were individually identified and sorted into 384-well plates. Whole genomes were amplified with multiple displacement amplification and the best amplification products based on real-time kinetics were sequenced to generate single-amplified genomes (SAGs). In total 59, 50, and 48 cellulose-microbe particles gave low crossing point (Cp) values (Supplementary Fig. 1) from time points 0, 1, and 2 months, respectively, and were sequenced yielding genomic information for a total survey of 157 cellulose-microbe particles