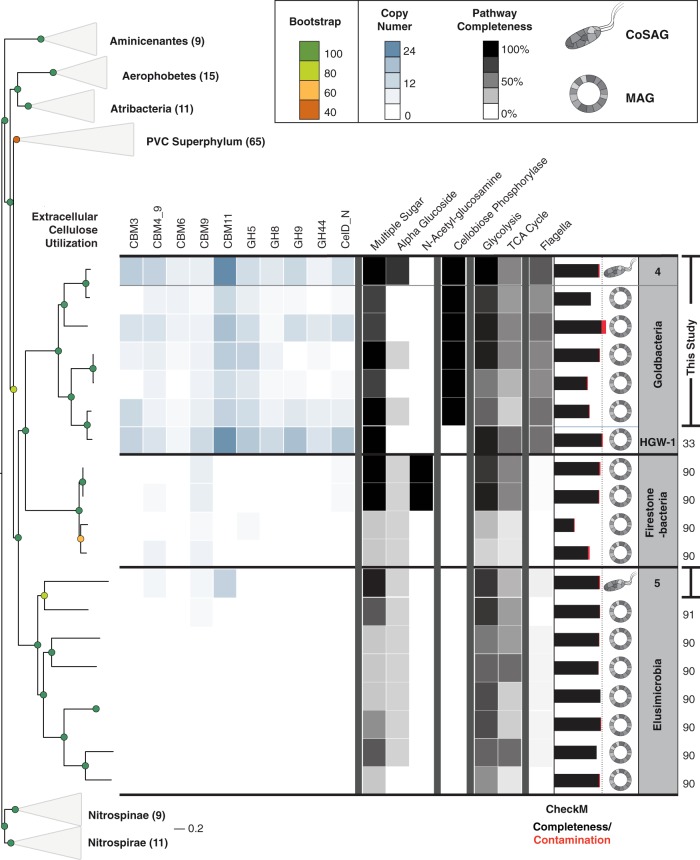
Fig. 4.
Characterization of our Goldbacteria genome (GBS-CoSAG_04) based on its placement in a single-copy marker gene tree together with its nearest neighbors: Firestonebacteria and Elusimicrobia, the additional MAGs extracted from published metagenomes ( Supplementary Table 2) and the previously published MAG, HGW-1 [41]. The heat map displays the families (Pfams) and KEGG Orthology (KO) terms associated with degradation, internalization, and central metabolism of cellulose as a substrate. Of note is the clear absence of cellulose degrading and flagellar genes in the Goldbacteria sibling clades, Firestonebacteria and Elusimicrobia. IMG metagenomes where MAGs were recovered are described in Supplementary Table 2. Brackets at far right indicate genomes derived from his study while numbers correspond to the following genomes/metagenomes available in the public databases [41, 86, 87]