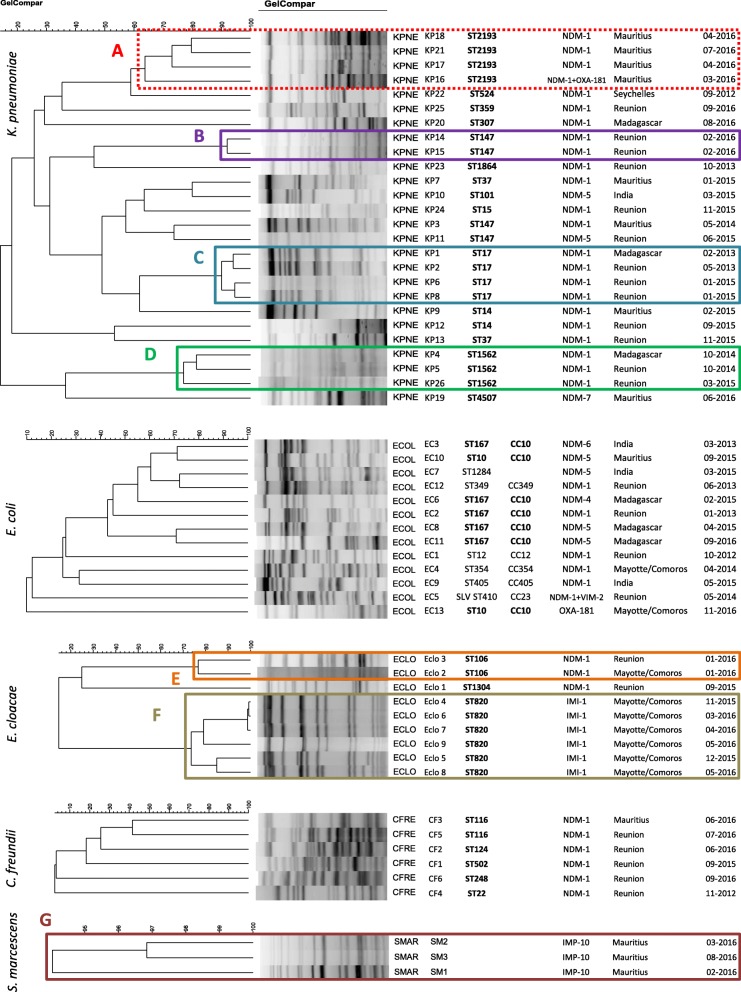
Fig. 2.
Molecular genotyping (PFGE and MLST analysis) applied to 57 CPE isolates of K. pneumoniae, E. coli, E. cloacae, C. freundii, and S. marcescens (2011–2016, Reunion Island, France). There is not MLST scheme definition for S. marcescens. Data presented: PFGE pattern, bacterial species (KPNE, K. pneumoniae; ECOL, E. coli; ECLO, E. cloacae; CFRE, C. freundii, SMAR, S. marcescens), sequence type, clonal complex, carbapenemase(s) produced, foreign country visited by the patient within the year before CPE isolation, month and year of CPE isolation. The similarity scales are specific of each bacterial species. The optimization and band-tolerance values used for PFGE dendrogram were 1%