Fig. 1 |. regulation of synaptic plasticity-related gene expression through epigenetic mechanisms.
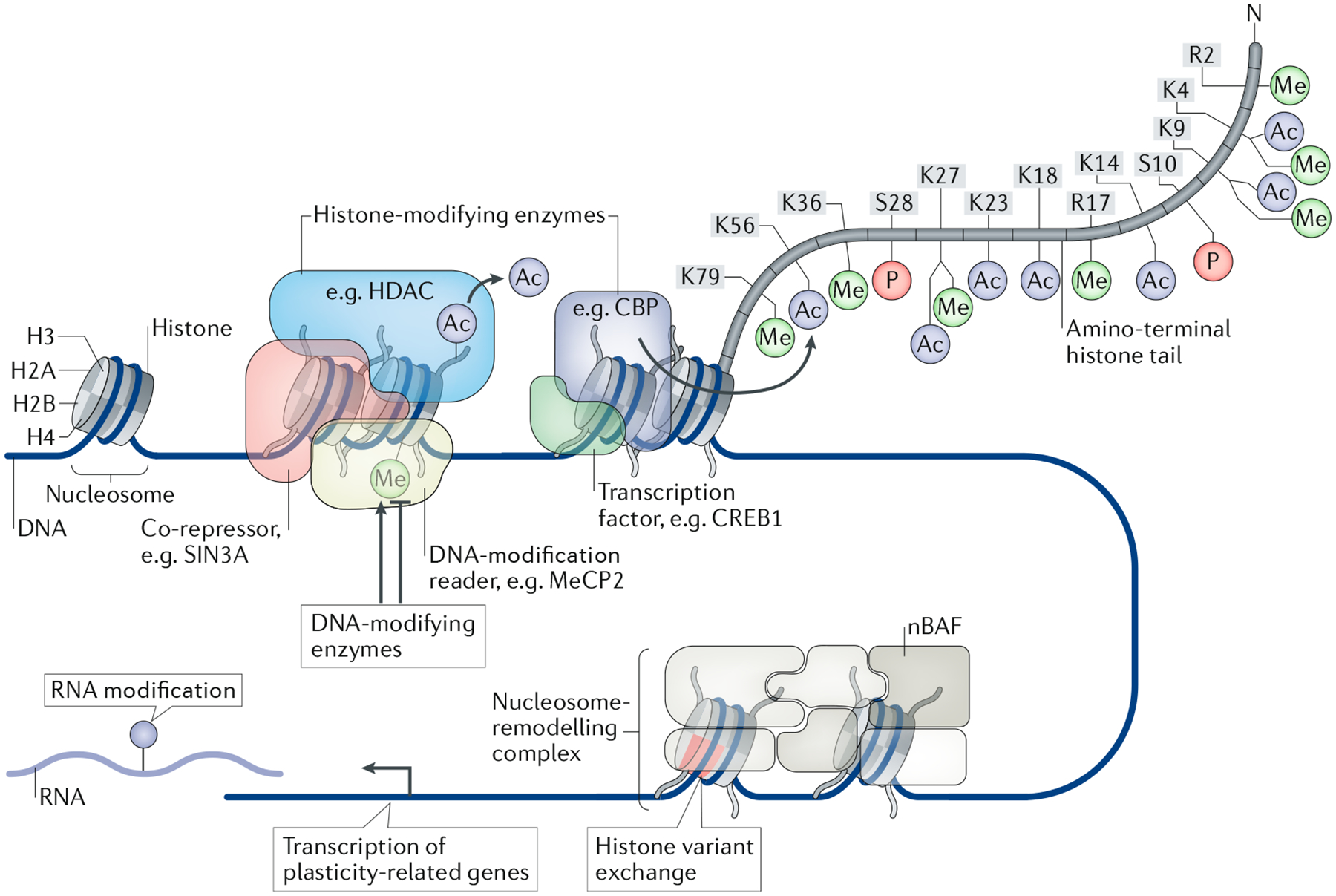
Several epigenetic mechanisms have been identified as regulators of gene expression important for synaptic plasticity and memory formation. For instance, histone acetylation mediated by the activity of histone acetyltransferases, such as CREB-binding protein (CBP), can facilitate memory-related gene expression. CBP is recruited by the transcription factor cAMP-responsive element-binding protein 1 (CREB1) and promotes a permissive transcription environment by adding acetyl groups onto the lysine tails of histones. By contrast, histone deacetylases (HDACs) remove acetyl groups from histone tails and act in concert with associated co-repressor transcription factors to reduce gene expression (for example, transcriptional co-repressor SIN3A). Gene expression can be repressed by the interaction with epigenetic enzymes, such as HDAC–repressor complexes or methyl-CpG-binding protein 2 (MeCP2), which binds to methylated DNA. DNA methylation is controlled by several DNA-modifying enzymes, including DNA methyltransferase 3A (DNMT3A) or DNMT3B and ten-eleven translocation enzymes (TETs), which reportedly repress or permit gene expression depending on the region of DNA that is methylated. Nucleosome-remodelling complexes, such as the neuronal BRG1-associated factor (nBAF) complex, interact with DNA and histones to potentially regulate chromatin structure and synapse-related gene expression through insertion of histone variants, nucleosome sliding, nucleosome eviction and chromatin looping. Although RNA-modifying enzymes do not directly affect chromatin structure, they do influence the rate of mRNA translation and the localization of RNAs, including at the synapse.
