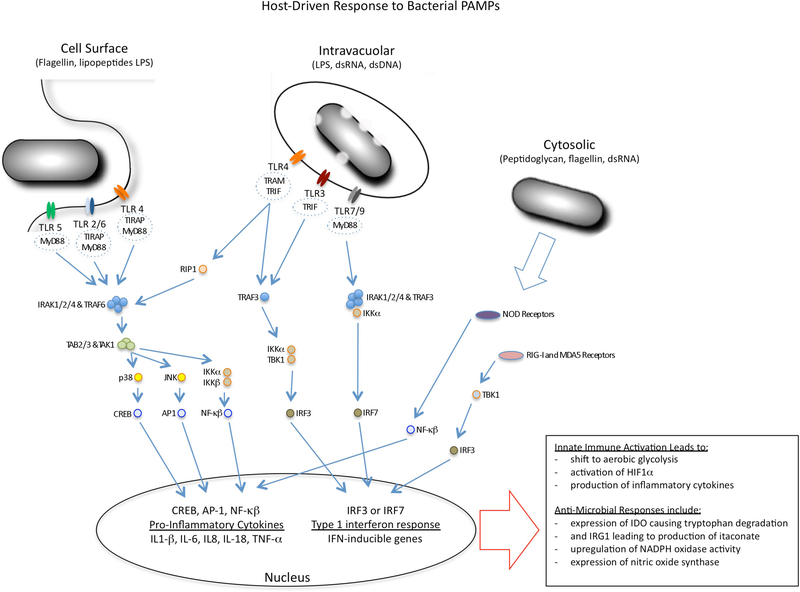
Figure 2 |. Modulation of macrophage metabolism by microbial mediators.
Microorganisms modulate the metabolic status of macrophages through the activation of innate immune signalling pathways or through the use of specific effectors, known as ‘virulence’ factors. The figure summarizes the major pattern recognition receptors (PRRs) found at the cell surface of macrophages and inside the cells, together with key components of their signalling pathways. Microorganisms activate Toll-like receptor (TLR) signalling pathways in host cells through their nucleic acids and cell wall constituents — known as pathogen-associated molecular patterns (PAMPs) — which drive macrophages into a state of enhanced glycolysis and increased production of lactate. The TLRs sense their ligands within the endosomal network as well as at the cell surface. They use different adaptor proteins that can modulate the outcome of TLR ligation, although the dominant responses are linked to either the production of pro-inflammatory cytokines through the activation of nuclear factor-κB (NF-κB) or the induction of a type I interferon response through interferon regulatory factor 3 (IRF3) and IRF7. PAMPs that are delivered into the cell cytosol by bacterial secretion systems activate signalling pathways through NOD-like receptors (NLRs), retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), with similar metabolic and cellular outcomes. The metabolic shift in the macrophages is accompanied by the upregulation of antimicrobial responses, including expression of indoleamine 2,3-dioxygenase (IDO) to limit tryptophan availability, production of itaconate through the activity of immunoresponsive gene 1 (IRG1) to intoxicate bacterial metabolism, and enhanced production of reactive oxygen and nitrogen species. ds, double-stranded; HIF1α, hypoxia inducible factor 1α; IKK, inhibitor of NF-κB kinase; IRAK, IL-1 receptor-associated kinase; LPS, lipopolysaccharide; MYD88, myeloid differentiation primary-response gene 88; NOS2, inducible nitric oxide synthase; TBK1, TANK-binding kinase 1; TIRAP, Toll/IL-1R (TIR)-domain-containing adaptor protein; TRAF, TNF receptor associated factor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain-containing adaptor protein inducing IFNβ.