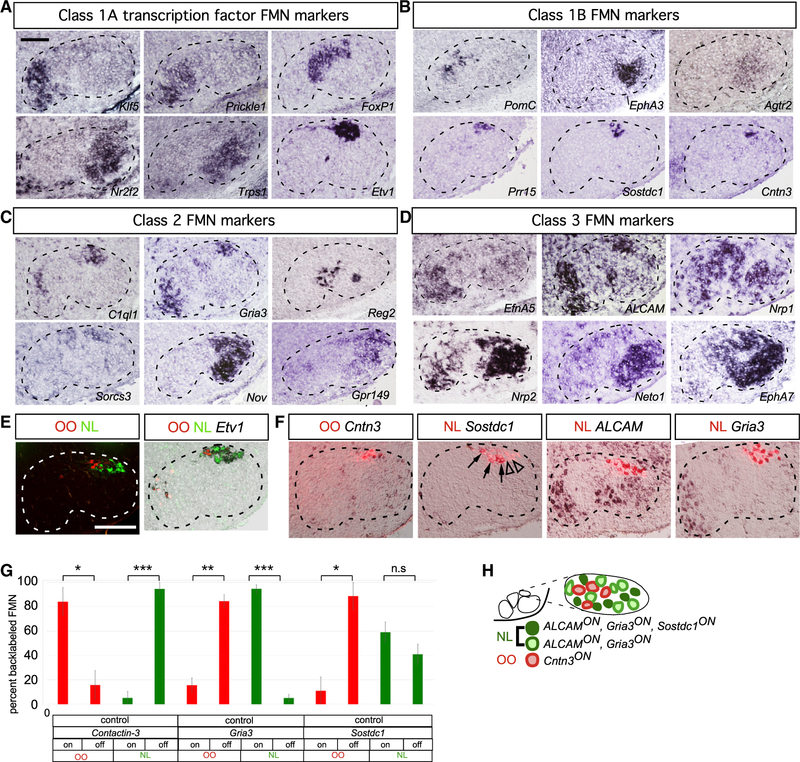
Figure 2. Unique, Combinatorial Gene Expression Programs Define Facial Motor Subnuclei and Pools.
(A–F) ISH for facial subnucleus markers in coronal cryosections at E16.5 (A–D) and P3 (E and F). Dashed lines in (A)–(F) outline the approximate boundary of the facial nucleus.
(A) Class 1A microarray hits defining distinct facial subnuclei.
(B) Class 1B microarray hits marking subpopulations within a single subnucleus.
(C) Class 2 microarray hits defining two subnuclei or subpopulations therein.
(D) Class 3 microarray hits marking FMNs of three or more facial subnuclei.
(E) Simultaneous injection of retrograde Alexa 555- and Alexa 647-conjugated cholera toxin B (A555CTB and A647CTB) labels into OO and NL muscles, respectively, marking distinct motor pools within the Etv1ON DL facial-motor subnucleus. Representative images.
(F) Single CTB injection followed by ISH identifies Cntn3 as an OO motor pool marker, whereas Sostdc1, ALCAM, and Gria3 define the NL pool. Sostdc1ON (arrows) and Sostdc1OFF (arrowheads) NL FMNs are indicated.
(G) Specificity of motor pool markers in (F) represented as mean ± SE percentage of FMNs marked by retrograde muscle labeling expressing the indicated motor-pool markers. n = 3 mice per muscle injection, (*p < 0.05, **p < 0.005, ***p < 0.0005, unpaired t test).
(H) Summary of OO and NL motor-pool molecular identities.
Scale bars: 100 μm in (A)–(D) and 200 μm (E) and (F).
See also Figures S1, S2, and S6.