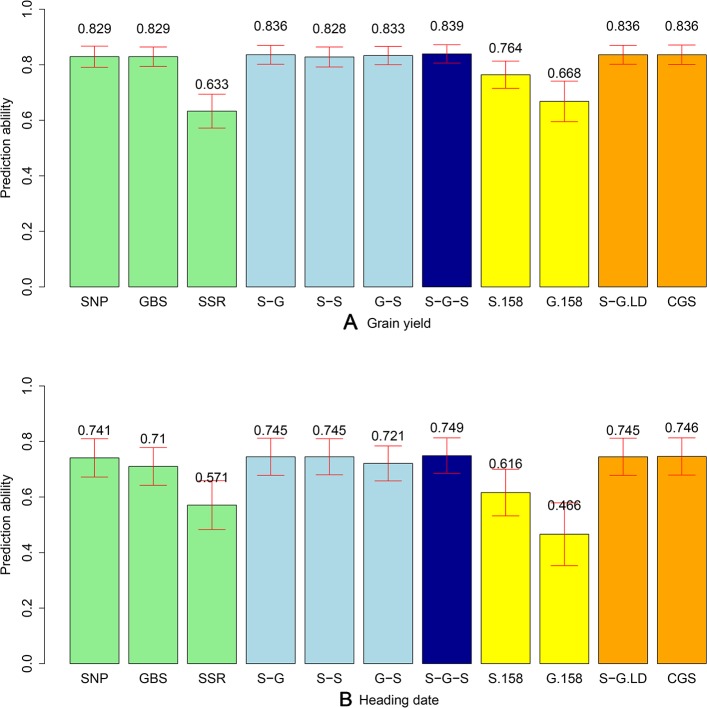
Figure 3.
Bar plot of average prediction abilities derived from 1,000 cross-validations from different prediction models for (A) grain yield and (B) heading date (HD). Single kernel models (green) were used for data from single-nucleotide polymorphism (SNP) array, genotyping-by-sequencing (GBS), and SSR markers. Double-kernel models (light blue) were used combining SNP array and GBS markers (S-G), SNP array and SSR markers (S-S), as well as GBS and SSR markers (G-S). The three-kernel model (dark blue) combined SNP array, GBS, and SSR markers (S-G-S). Subsets of 158 markers from SNP array markers (S.158) and GBS markers (G.158) were used to run the single kernel models (yellow). Moreover, after ignoring the GBS markers with higher linkage equilibrium with SNP array markers, a double-kernel model combing SNP array and remained GBS markers (S-G.LD) and a single-kernel model of the combination of SNP array and remained GBS markers (CGS) (orange) were used. The corresponding standard deviations are illustrated as red bars.