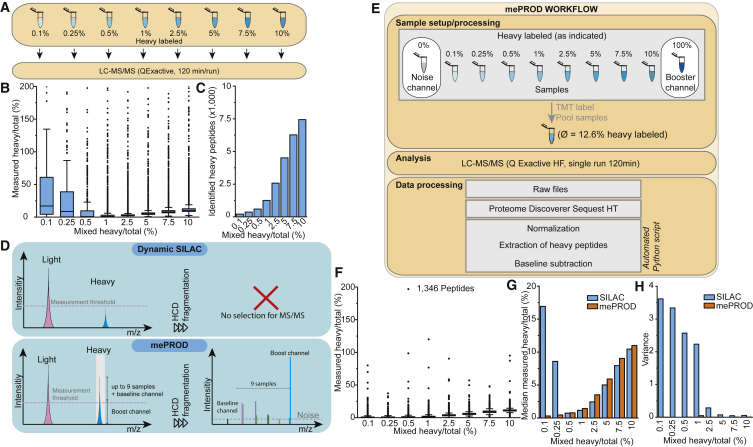
Figure 1.
mePROD Proteomics Overcomes Low Accuracy and Identifications of Peptides at Low Heavy-to-Light Ratio
(A) Scheme of experimental design. Heavy and light peptides were mixed at indicated ratios.
(B) Measured heavy to total ratios on peptide level. Boxes indicate 25%/50% quartiles and the median; whiskers show standard deviation.
(C) Number of heavy labeled peptides quantified in (B).
(D) Underlying principle of mePROD to increase signals of interest. Low labeling stoichiometry prevents reaching the measurement threshold using standard dynamic SILAC approaches (top). In mePROD, a booster channel comprised of a fully heavy labeled proteome boosts the signal of interest above the MS1 detection level (bottom). Heavy/total ratios for individual samples are then then determined from TMT signals quantified in MS2 (right).
(E) Experimental mePROD design and data processing. Samples from (A) were combined with noise and booster channels, TMT-labeled, pooled, analyzed by LC-MS2, and raw files processed. Reporter ion intensities for peptides were sum normalized and heavy peptide intensities extracted. To enhance accuracy, baseline values derived from the non-SILAC labeled channel were subtracted from each peptide.
(F) Samples as in (A) were analyzed using mePROD (using 1/8th of the LC-MS2 machine time used in A). Comparison of measured versus expected heavy/total ratios. Boxes indicate 25%/50% quartiles and the median; whiskers show standard deviation.
(G and H) Comparison of median measured heavy/total peptide ratios (G) or variance (H) for samples measured by SILAC or mePROD. See also Figure S1.