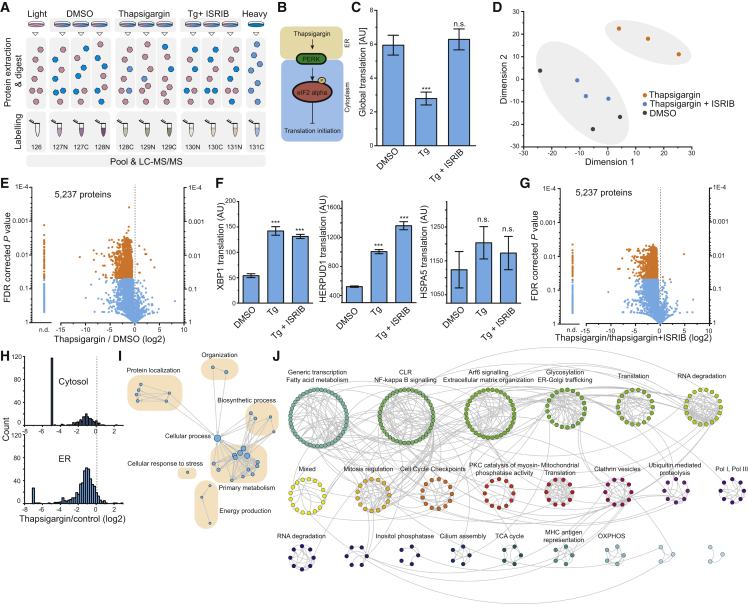
Figure 3.
Changes in the Cellular Translatome upon Activation of the Integrated Stress Response by Protein Misfolding in the Endoplasmic Reticulum
(A) Experimental layout. Three different conditions were pooled (in triplicate) with noise and booster channels and analyzed by mePROD MS.
(B) Scheme of translational repression during the UPR, induced by PERK activation.
(C and D) Global translation levels assessed by mePROD MS for cell treated with DMSO, 1 μM thapsigargin (Tg), or 1 μM thapsigargin and 500 nM ISRIB (Tg + ISRIB) for 2.5 h. Shown are median intensities of heavy labeled peptides (C). Error bars indicate standard deviation (n = 3). ∗∗∗p < 0.001; n.s., not significant (two-sided, unpaired Student’s t test with equal variance). AU, arbitrary units. Multidimensional scaling analysis of samples standardized by unit variance (D).
(E) Volcano plot showing fold change of relative translation versus adjusted p value of thapsigargin versus control treated cells. Orange dots indicate significantly changing proteins (p values < 0.05 and fold change [log2] ≤ −0.5 or ≥ 0.5). Samples for which abundances in thapsigargin treated samples dropped below baseline and no fold change could be calculated are indicated as not determinable (n.d.).
(F) Changes in translation levels of XBP1, HERPUD1, and HSPA5 (better known as BIP) measured by mePROD MS. Mean heavy abundance was plotted with error bars indicating standard deviation (n = 3). ∗∗∗p < 0.001; n.s., not significant (two-sided, unpaired Student’s t test with equal variance). Tg, thapsigargin.
(G) Volcano plot showing fold change versus adjusted p value between thapsigargin and thapsigargin+ISRIB treated samples. Significantly changing proteins in orange (as in E).
(H) Histogram depicting translation changes for cytosolic versus endoplasmic reticulum resident proteins.
(I) EnrichmentMap network showing significantly (q value < 0.001) enriched GO terms for proteins without significantly changed relative translation rates upon thapsigargin treatment.
(J) ReactomeFI cluster analysis for proteins not changing relative translation rates upon thapsigargin treatment. Proteins were FI annotated, clustered, and clusters analyzed for significantly enriched Reactome pathways (q value < 0.001). The most prominent pathway of each cluster is indicated. Connecting lines show interaction of protein nodes. See also Table S1 and Figures S2–S4.