Figure 1.
RING1B I53S Is Fully Catalytically Dead In Vivo
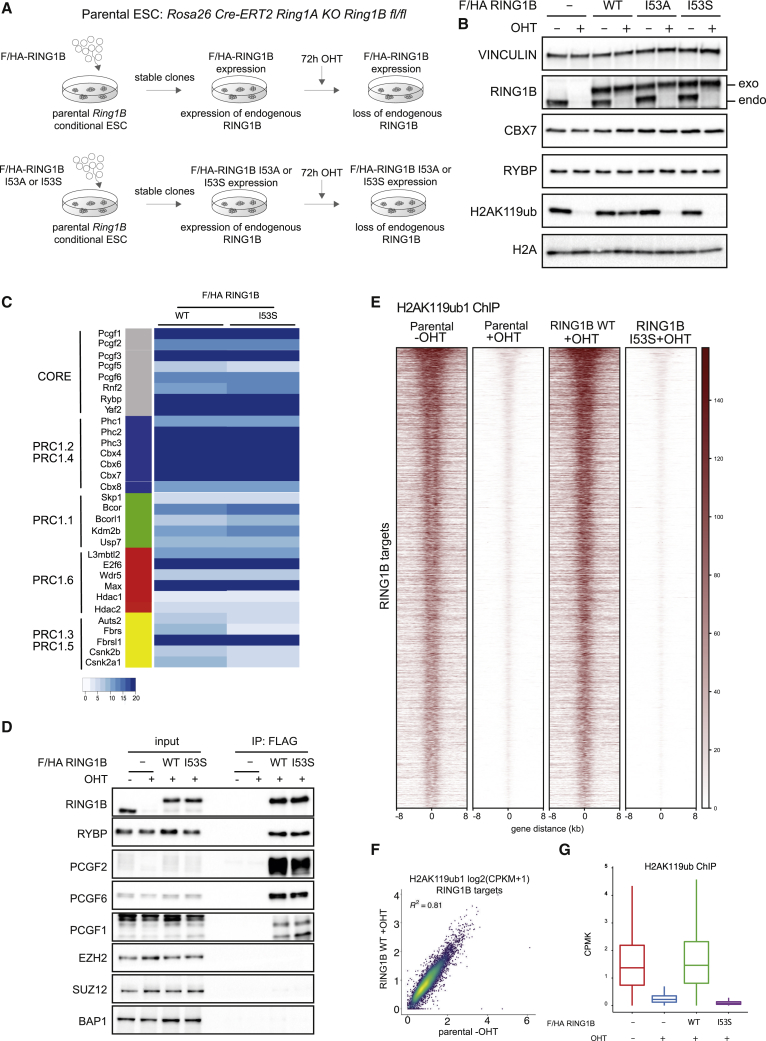
(A) Schematic representation of the strategy used for the generation of ROSA26::creERT2 RING1A−/−;RING1Bfl/fl conditional mESCs stably expressing FLAG-HA (F/HA)-tagged RING1B WT or I53A/S.
(B) Western blot analysis with the indicated antibodies of total protein extracts obtained from the specified cell lines upon 72 h of treatment with OHT (+OHT) or EtOH (−OHT). Vinculin and histone H2A were used as loading controls.
(C) Values of the LFQ ratios of the RING1B WT and I53S obtained by tandem mass spectrometry (MS/MS) analyses in the RING1B immuno-purifications (anti-FLAG) from ROSA26::creERT2 RING1A−/−;RING1Bfl/fl conditional mESCs stably expressing FLAG-HA (F/HA)-tagged RING1B WT or I53S upon 72 h of treatment with OHT (+OHT).
(D) Co-immunoprecipitation analysis of nuclear extracts derived from FLAG-HA (F/HA)-tagged RING1B WT or I53S expressing cells upon 72 h of treatment with OHT (+OHT) or EtOH (−OHT) using M2 affinity gel beads. FLAG-IPs in parental cells served as a negative control.
(E) Heatmaps representing normalized H2AK119ub1 ChIP-seq intensities ±8 kb around the center of RING1B target loci in the indicated cell lines.
(F) Scatterplot showing the relationship between H2AK119ub1 CPMK levels (counts per million per kilobase) between parental EtOH treated (−OHT) and RING1B WT OHT-treated (+OHT) cells in RING1B target loci. R2 represents the coefficient of determination of linear regression.
(G) Boxplots representing H2AK119ub1 ChIP-seq CPMK levels in the indicated cell lines at RING1B target loci.
See also Figure S1A.