Figure 2.
The Plant-specific N-terminal Extension of AtTSPO Is Required for Interaction and Depletion of PIP2;7 In Vivo
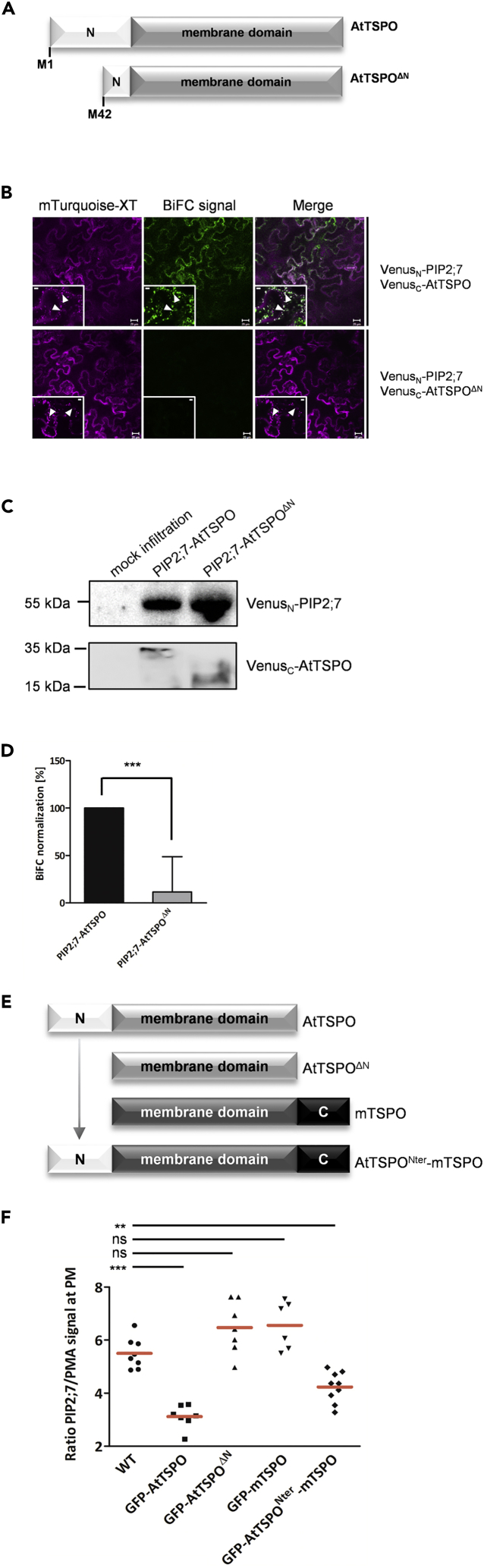
(A) Schematic representation of AtTSPO genetic constructs prepared for BiFC analysis. Full-length PIP2;7 and AtTSPO served as positive controls. The N-terminally truncated variant starts with methionine at position 42.
(B) Representative confocal images of tobacco epidermal cells transiently coexpressing VenusN-PIP2;7 and VenusC-AtTSPO or VenusN-PIP2;7 and VenusC-AtTSPOΔN. Xylosyltransferase-mTurquoise fluorescent chimera (magenta) was imaged as a cell transfection control (Golgi marker) and for signal quantification. Low-magnification images qualitatively demonstrate the occurrence or lack of BiFC (green), and insets of high magnification images show Golgi stacks (arrowheads). Bars = 20 μm and 5 μm for low and high magnification, respectively. Experiments were repeated three times.
(C) Western blot of total extracts from infiltrated leaf areas imaged by BiFC. PIP2;7 expression was detected by anti-GFP. Full-length and truncated AtTSPO were detected by anti-FLAG (tag originally cloned between VenusC and AtTSPO). Non-infiltrated leaf area served as a negative control.
(D) Signal quantification shows a drastic reduction in BiFC, and hence interaction between PIP2;7 and truncated AtTSPO mutant (gray histogram) compared with full-length AtTSPO (black histogram). Bars represent means +/− SD. Statistical significance was assessed by an independent samples t test using Graphpad Prism (***p < 0.001).
(E) Schematic representation of genetic constructs stably expressed in Arabidopsis suspension cells. The function of the AtTSPO N-terminus was assessed using the mouse homolog (mTSPO) as a negative control and carrier protein for the plant N-terminus. Each construct was N-terminally tagged with GFP.
(F) PM fractions were extracted from Arabidopsis suspension cells and PIP2;7 was quantified by western blot. PM proton ATPase (PMA) served as a reference protein for signal quantification. Individual measurements (PIP2;7 signal normalized against PMA signal from 6 to 9 replicates from two independent PM preparations) are shown, and the red horizontal lines define the means. Statistical significance was analyzed by t-tests (D) or one-way ANOVA followed by Tukey's tests (**p < 0.01; ***p < 0.001; ns, not significant) using Graphpad Prism.