Figure 1.
RING1BI53A/D56K Is Catalytically Inactive but Not Sufficient to Eliminate H2AK119ub1 In Vivo
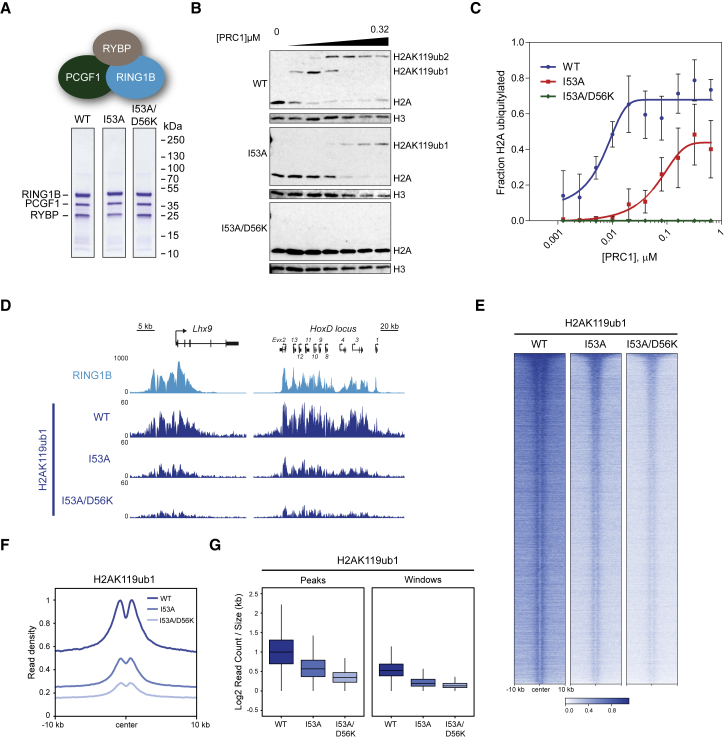
(A) Coomassie-stained gels of affinity-purified RING1B-PCGF1-RYBP complexes.
(B) In vitro E3 ubiquitin ligase assays in which conversion of unmodified histone H2A to ubiquitylated forms was measured by western blot with an H2A-specific antibody. An H3-specific antibody was used as a control.
(C) Quantification of the mean fraction of histone H2A ubiquitylation across a range of PRC1 concentrations from in vitro E3 ubiquitin ligase assays in (B). Error bars show SEM (n = 2 or more).
(D) Genomic snapshots of classical RING1B-bound loci, showing cChIP-seq for RING1B in wild-type ESCs and cChIP-seq for H2AK119ub1 in RING1BWT, RING1BI53A, and RING1BI53A/D56K ESCs.
(E) Heatmap analysis of H2AK119ub1 cChIP-seq at RING1B-bound sites in RING1BWT, RING1BI53A, and RING1BI53A/D56K ESCs. Genomic regions were sorted based on RING1B occupancy in untreated PRC1CKO ESCs.
(F) Metaplot analysis of data shown in (E).
(G) Boxplots comparing normalized H2AK119ub1 cChIP-seq signal at RING1B-bound sites and 100 kb genomic windows in RING1BWT, RING1BI53A, and RING1BI53A/D56K ESCs.
See also Figure S1.