Figure 3.
PRC2 Binding and H3K27me3 Deposition at Polycomb Target Sites Rely on PRC1 Catalytic Activity
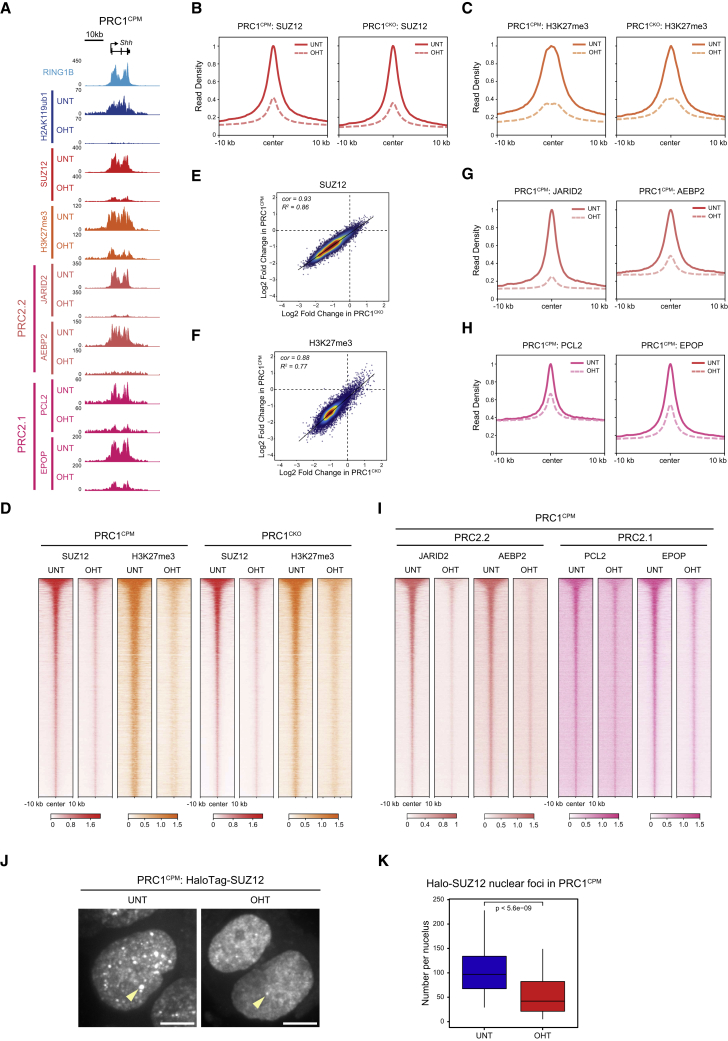
(A) Genomic snapshot of a Polycomb target gene showing cChIP-seq for RING1B, H2AK119ub1, SUZ12, H3K27me3, JARID2, AEBP2, PCL2, and EPOP in PRC1CPM cells.
(B) Metaplot analysis of SUZ12 cChIP-seq at PcG-occupied sites in PRC1CPM and PRC1CKO cells.
(C) As in (B) for H3K27me3 cChIP-seq.
(D) Heatmap analysis of cChIP-seq data shown in (B) and (C). Genomic regions were sorted based on RING1B occupancy in untreated PRC1CKO ESCs.
(E) A scatterplot comparing the log2 fold changes in SUZ12 levels at PcG-occupied sites in PRC1CPM and PRC1CKO ESCs. R2 represents coefficient of determination for linear regression and cor denotes Pearson correlation coefficient.
(F) As in (E) for H3K27me3 cChIP-seq.
(G) Metaplot analysis of PRC2.2-specific subunits JARID2 and AEBP2 cChIP-seq at PcG-occupied sites in PRC1CPM cells.
(H) As in (G) for PRC2.1-specific subunits PCL2 and EPOP.
(I) Heatmap analysis of cChIP-seq data shown in (G) and (H). Genomic regions were sorted as in (D).
(J) Maximum intensity projections of JF549-Halo-SUZ12 signal in PRC1CPM ESCs. Examples of SUZ12 nuclear foci (Polycomb bodies) are indicated by arrowheads. Scale bar is 5 μm.
(K) Boxplots comparing the number of JF549-Halo-SUZ12 nuclear foci in PRC1CPM ESCs before (ncells = 55) and after (ncells = 52) OHT treatment. Cells from two independent experiments were analyzed. P values denote statistical significance calculated by a Student’s t test.
See also Figure S3.