Figure 4.
PRC1 Catalytic Activity Drives Canonical PRC1 Occupancy and Long-Range Polycomb Chromatin Domain Interactions
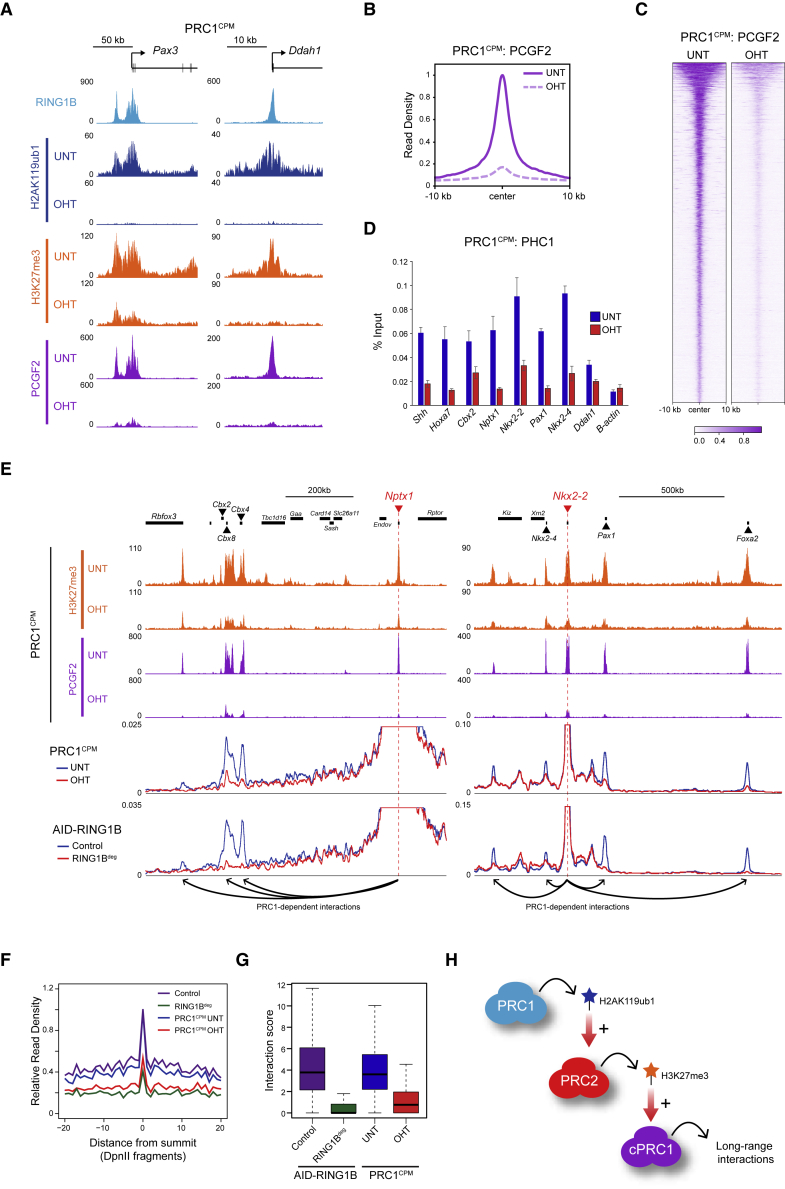
(A) Genomic snapshots of Polycomb target genes, showing cChIP-seq for RING1B, H2AK119ub1, H3K27me3, and PCGF2 (cPRC1) in PRC1CPM cells.
(B) Metaplot analysis of PCGF2 cChIP-seq at PCGF2 target sites in PRC1CPM cells.
(C) Heatmap analysis of data shown in (B). Genomic regions were sorted based on RING1B occupancy in untreated PRC1CKO ESCs.
(D) ChIP-qPCR for PHC1 at a panel of Polycomb target genes in PRC1CPM cells. B-actin is an active gene not bound by PHC1. Error bars show SEM (n = 3).
(E) Genomic snapshots of two regions encompassing classical Polycomb target sites used as baits in CaptureC (bait probe positions are marked by dashed red lines). H3K27me3 and PCGF2 cChIP-seq data in PRC1CPM cells are shown at the top. Below is the bait interaction landscape measured by CaptureC in PRC1CPM cells (untreated and OHT-treated) and auxin-treated Ring1a−/−;AID-RING1B (RING1Bdeg) ESCs (relative to the Control cell line with intact PRC1). Arrows illustrate long-range PRC1-dependent interactions.
(F) Meta-analysis of interactions between bait Polycomb target regions and other PCGF2 target sites (n = 130) in cells described in (E). Read density is shown relative to the untreated PRC1CPM cells.
(G) Boxplot analysis of CHiCAGO scores for the interactions between the bait Polycomb target regions and other PCGF2 target sites (n = 130) for cells described in (E).
(H) A schematic summarizing the model in which deposition of H2AK119ub1 by PRC1 drives accumulation of PRC2 and deposition of H3K27me3 at target sites, promoting cPRC1 binding and long-range interactions between these regions.
See also Figure S4.