Figure 5.
Variant PRC1 Complexes Occupy Polycomb Target Sites Independently of PRC1 Catalysis
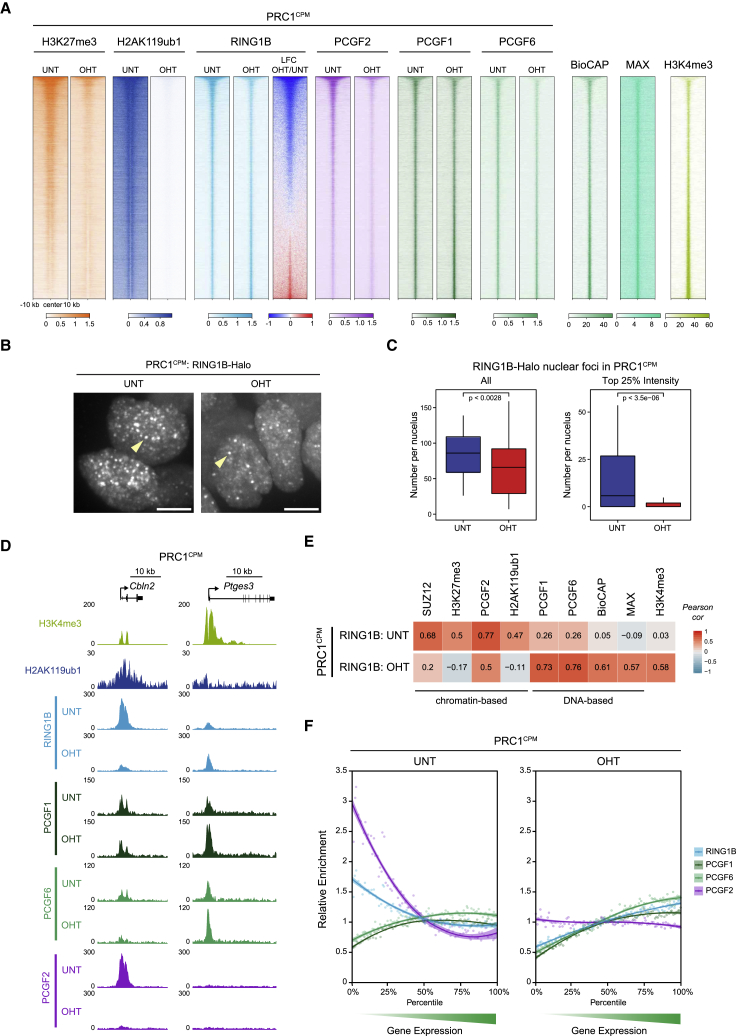
(A) Heatmap analysis of cChIP-seq for H3K27me3, H2AK119ub1, RING1B, PCGF2, PCGF1, and PCGF6 at RING1B-bound sites in PRC1CPM cells. Also shown is BioCAP-seq (measure of non-methylated CpG-rich DNA) and ChIP-seq for MAX and H3K4me3 in wild-type ESCs. Genomic intervals were sorted based on log2 fold change in RING1B occupancy following OHT treatment in PRC1CPM ESCs.
(B) Maximum intensity projections of RING1B-Halo-JF549 signal in PRC1CPM ESCs. Examples of RING1B nuclear foci are indicated by arrowheads. Scale bar is 5 μm.
(C) Boxplots comparing the number of all (left panel) or top 25% highest intensity (right panel) RING1B-Halo-JF549 foci per nucleus in PRC1CPM ESCs before (ncells = 69) and after (ncells = 83) OHT treatment. Cells from two independent experiments were analyzed. P values denote statistical significance calculated by a Student’s t test.
(D) Genomic snapshots of two genes showing cChIP-seq for RING1B, PCGF1, PCGF6, and PCGF2 in PRC1CPM ESCs. H2AK119ub1 cChIP-seq in untreated cells and H3K4me3 ChIP-seq in wild-type ESCs is also shown. Cbln2 is a lowly transcribed gene with high-level RING1B occupancy and Ptges3 is a more highly transcribed gene.
(E) Correlation of cChIP-seq signal for RING1B with PRC2 (SUZ12 and H3K27me3) and PRC1 (PCGF2, PCGF1, PCGF6, and H2AK119ub1) in untreated and OHT-treated PRC1CPM ESCs. Correlation with BioCAP-seq and ChIP-seq for MAX and H3K4me3 in wild-type ESCs is also shown.
(F) Relative enrichment of RING1B, PCGF1, PCGF6, and PCGF2 cChIP-seq signal in PRC1CPM ESCs across promoter-proximal RING1B-bound sites divided into percentiles based on the expression level of the associated gene in untreated PRC1CPM cells. For each factor, enrichment is shown relative to the fiftieth percentile. Lines represent smoothed conditional means based on loess local regression fitting.
See also Figure S5.