Figure 6.
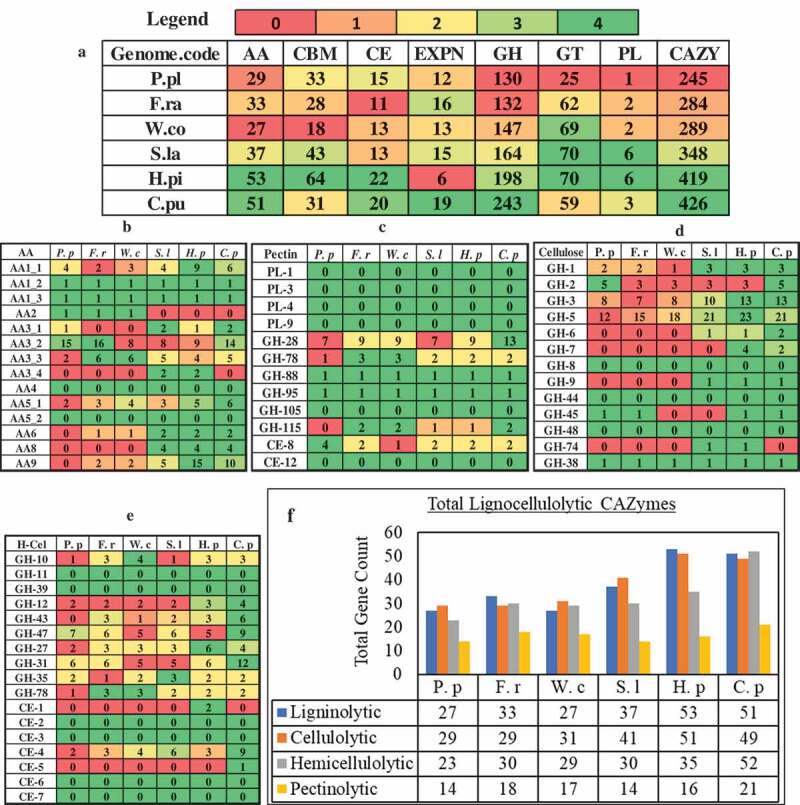
Pictorial representation of genome-wide CAZymes (a) genome wide distribution of CAZymes among all the selected brown rot fungi. (b) ligninolytic- auxiliary activity (AA), (c) pectinolytic, (d) cellulolytic, (e) hemicellulolytic, (f) tentative distribution of genes encoding for lignocellulolytic CAZymes in Rhodonia placenta (P. p), Fibroporia radiculosa (F. r), Wolfiporia cocos (W. c), Coniophora puteana (C. p), Serpula lacrymans (S. l), Hydnomerulius pinastri (H. p). [Note: Figure 5(a): AA = Auxiliary Activity, CBM = Carbohydrate binding modules, CE = Carbohydrate esterases, EXPN = Expansins, GH = Glycoside hydrolases, PL = Polysaccharide lyases and CAZY = Total number of carbohydrate active enzymes] .
