Fig. 3.
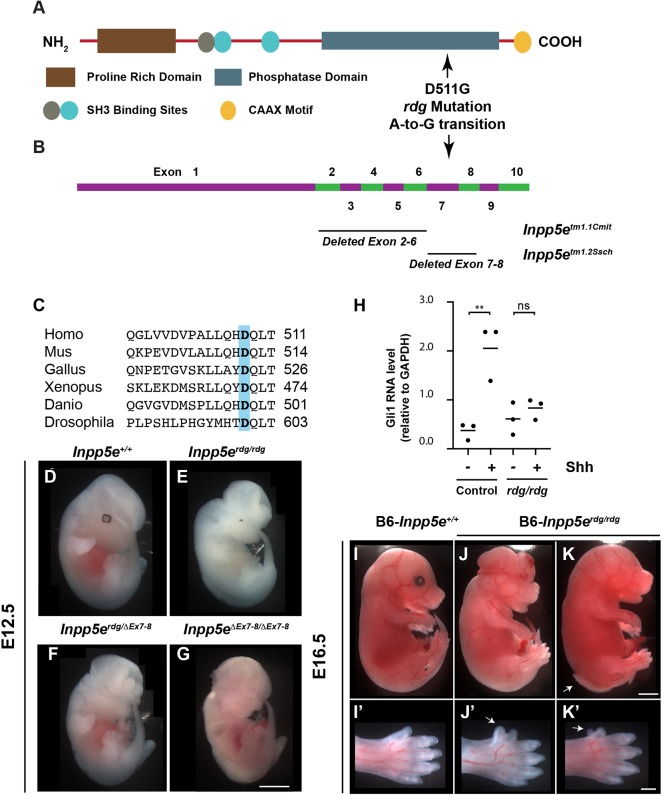
Inpp5erdg, which carries a D511G mutation, is a functional null allele of Inpp5e. (A) Schematic showing the protein domain structure of Inpp5e. D511G in Inpp5erdg is the result of an A-to-G transition in the coding strand (NM_033134.3:c2164A>G). (B) Exon structure of Inpp5e, based on NCBI reference sequence NM_033134.3, showing location of deleted exons in two previously characterized alleles. (C) Alignment of protein sequence surrounding position 511 of mouse Inpp5e (highlighted) showing that aspartic acid is conserved across multiple species. (D-G) Whole-mount images of E12.5 embryos. (E) Inpp5erdg/rdg mutants display exencephaly and microphthalmia (n=20). These animals are on a mixed FVB/C3H background so that the retinal pigment epithelium cells are visible. (F,G) Inpp5erdg/ΔEx7-8 (n=4) and FVB-Inpp5eΔEx7-8/ΔEx7-8 (n=9) embryos resemble Inpp5erdg/rdg mutants, indicating the alleles fail to complement. (H) qRT-PCR of Gli1 in Inpp5erdg/+ (n=3) and Inpp5erdg/rdg (n=3) MEFs in the presence (+) and absence (−) of Shh treatment. Bar indicates mean of biological replicates with individual data points shown, analyzed by ANOVA using Tukey's correction for multiple comparisons. **P<0.01; ns, not significant. (I-K) Inpp5erdg/rdg mutants on C57BL/6J background survive to E16.5 and display exencephaly, microphthalmia and spina bifida to varying degrees of severity (n=6). Arrow indicates spina bifida. (I′-K′) Close-up images showing hindlimb preaxial polydactyly. Arrow indicates extra digit. Scale bars: 1 mm for D-K; 500 µm for I′-K′.