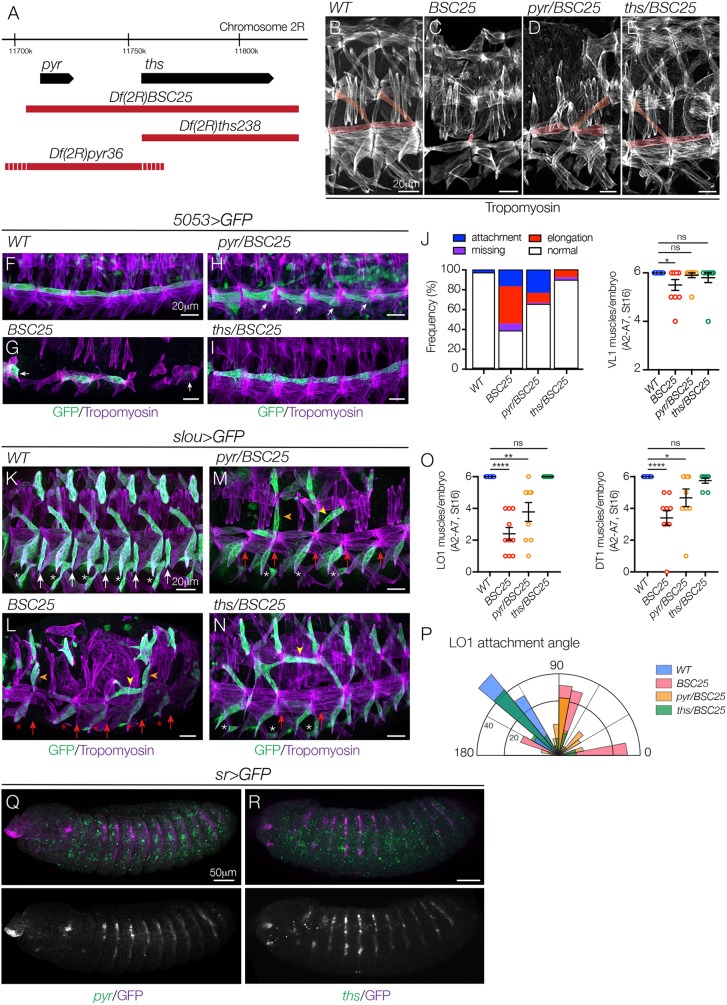
Fig. 4.
The FGF ligands Pyr and Ths direct myotube guidance. (A) Genomic organization of pyr and ths. Sequences deleted by Df(2R)BSC25, Df(2R)pyr36 and Df(2R)ths238 are shown in red (dashes reflect breakpoint uncertainty). Df(2R)pyr36 does not disrupt ths transcript expression (Kadam et al., 2009). (B-E) Stage 16 embryos labeled for Tropomyosin. Df(2R)BSC25 (C) and Df(2R)pyr36 embryos (D) showed severe body wall muscle defects, including missing and disorganized muscles, compared with WT (B). Df(2R)ths238 embryos (E) showed mild myogenic defects. VL1 and LO1 muscles are pseudocolored. (F-I) Stage 16 5053>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). VL1 muscles in Df(2R)BSC25 (G) and Df(2R)pyr36 embryos (H) showed elongation and attachment site defects, compared with WT (F). VL1 muscles in Df(2R)ths238 embryos were largely normal (I). White arrows show incorrect attachment sites. (J) Quantification of VL1 muscle phenotypes in stage 16 embryos (n≥54 muscles per genotype). Left shows a histogram of muscle phenotypes. Right shows the number of VL1 muscles. (K-N) Stage 16 slou>eGFP embryos labeled for GFP (green) and Tropomyosin (violet). LO1 muscles in Df(2R)BSC25 (L) and Df(2R)pyr36 embryos (M) acquired lateral (orange arrowheads) and transverse (yellow arrowheads) morphologies similar to htlAB42 LO1 muscles, compared with WT (K). LO1 muscles in Df(2R)ths238 embryos also showed morphological defects (N). White asterisks, VA3 muscles; red asterisks, missing VA3 muscles; white arrows, VT1 muscles; red arrows, missing VT1 muscles. (O) Quantification of LO1 (left) and DT1 (right) muscles. (P) Radial density plot of LO1 muscle attachment angles in stage 16 embryos (n≥35 muscles per genotype). (Q,R) Stage 12 sr>eGFP embryos labeled for pyr (Q) or ths (R) mRNA (green) and GFP (violet). pyr is expressed in the three ectodermal domains per segment and is excluded from Sr+ tendon cells (Q). ths expression in the ectoderm is modest but detectable immediately anterior to tendon cells (R). ns, not significant. *P<0.05, **P<0.01, ****P<0.0001 (unpaired, two-tailed Student's t-test). Data are mean±s.e.m. See also Fig. S2. Embryos are oriented with anterior to the left and dorsal to the top.