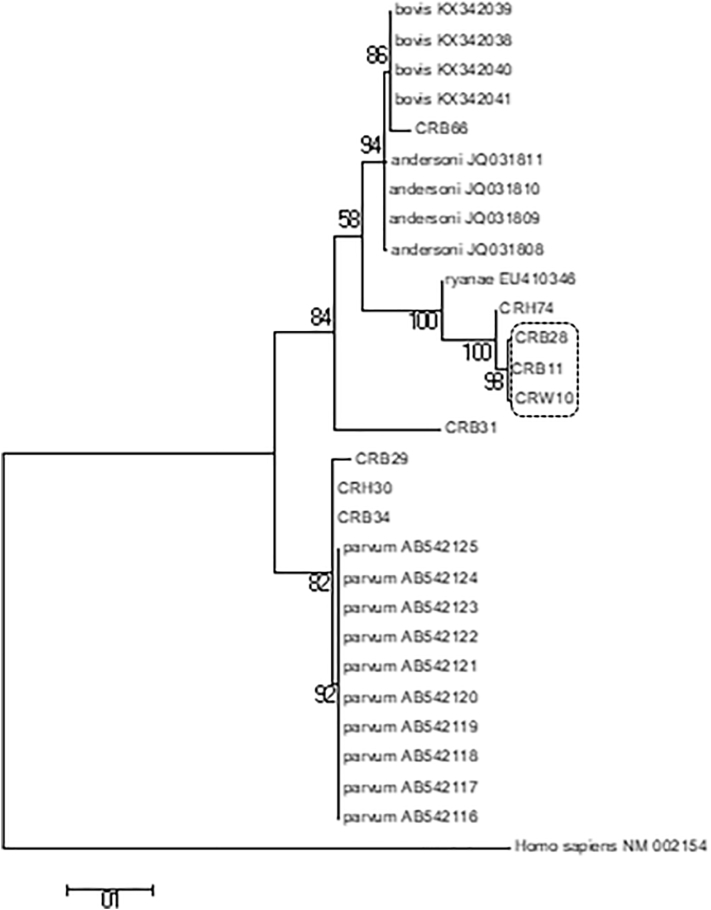
Fig. 1.
Phylogenetic analysis of our study isolates based on hsp70 gene. Previously published hsp70 sequences from C. parvum, C.ryanae C. andersoni and C. bovis were aligned with the representative sequences of our study isolates (a total of 9 sequences) using the ClustalW multiple alignment program of MEGA version 7 software. The phylogenetic tree was constructed from this alignment using maximum likelihood matrix algorithm. The bootstrap values were also analyzed to estimate confidence intervals. The study isolates from bovine and water samples formed a distinct cluster with high bootstrap value, which was marked with “dotted block”.