ABSTRACT
To investigate whether introduction of colistin into the clinical settings selected colistin-resistant CRE, we performed molecular epidemiological study of 1868 CRE strains collected from different geographical locales in China during the period 2014–2019. 1755 (96.18%) isolates carried the carbapenemase genes blaKPC and blaNDM; 14 Escherichia coli isolates (0.75%) carrying mcr-1 and blaNDM (MCR-CREC) were also identified. Importantly, the number and relative prevalence of MCR-CREC isolates increased from 5 (0.41%) to 9 (1.38%) after introduction of polymyxin into clinical practice. Consistently, results of genetic analysis indicated that MCR-CREC strains collected before December 2017 were genetically diverse, yet those collected after that date exhibited more closely related genetic profiles, indicating that specific MCR-CREC strains were rapidly selected as a result of increased usage of colistin in clinical settings. The resistance level of MCR-CREC isolates to colistin increased after the introduction of polymyxin into clinical use with the MIC to colistin from <2 mg/L in 80% strains to 2 mg/L in 100% strains. Further dissemination of MCR-CREC strains, which exhibit resistance to the last-line drugs of carbapenems and colistin, is expected to pose a severe threat to human health.
KEYWORDS: Carbapenem-resistant Enterobacteriaceae, mcr-1, colistin, clinical uses, China
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) strains pose a serious public health threat worldwide and are associated with high mortality rates [1,2]. Escherichia coli and Klebsiella pneumoniae are among the most frequently isolated CRE in China. In 2014, the carbapenem resistance rate of these two species was 1.0% and 13.4%, respectively [3,4]. The major mechanisms of carbapenem resistance in CRE include production of carbapenemase, hyper-production of the AmpC β-lactamase, and the combined effect of extended spectrum β-lactamases (ESBLs), porin impermeability, and over-expression of efflux pumps [5]. Carbapenemases commonly produced Enterobacteriaceae include KPC, NDM, VIM, IMP, and OXA-48 [6]. A nationwide surveillance of CRE strains conducted in China indicated that the major carbapenemases produced by carbapenem-resistant K. pneumoniae (CRKp) and carbapenem-resistant E. coli (CREc) were KPC (74%) and NDM (49%), respectively, and that the predominant sequence types of CRKp and CREc were ST11 and ST131/ST167, respectively [2,7].
Current treatment options for infections caused by CRE are severely limited. Two β-lactam/β-lactamase inhibitor combinations, ceftazidime/avibactam and ceftolozane/tazobactam, approved for treatment of multidrug resistant gram-negative bacteria abroad, were not available for clinical use in China until June 2019 when ceftazidime–avibactam was officially approved [8]. Treatment regimens against CRE typically rely on last-line antibiotics such as tigecycline and polymyxin [9]. Polymyxin was re-classified as a “critically important” human medicine by WHO in 2012 [10]. In China, polymyxin was approved for use as an injection drug in treatment of bacterial infections in January 2017, and was adopted for clinical use in late 2017 (http://www.mohrss.gov.cn/gkml/zcfg/gfxwj/201702/t20170223_266775.html). Increased usage of polymyxins in clinical settings, however, has led to the emergence of polymyxin-resistant CRE in China and various countries worldwide, especially those carrying the plasmid-borne mcr variants [11,12]. In an effort to control polymyxin resistance, the Ministry of Agriculture of China (article number 2428) withdrew colistin from the list of feed additives and growth promoters in November 2016, and this policy was officially enforced in April 2017 [13].
Polymyxin E, also known as colistin, is an important cationic antimicrobial peptide whose clinical potential has been significantly compromised by the global spread of the plasmid-borne colistin resistance genes mcr [9,14]. MCR-1 is a phosphoethanolamine transferase firstly reported in late 2015 [14]. A Chinese nationwide epidemiological study revealed a fairly high prevalence of MCR-1-positve E. coli among humans (3.7–32.7% among different provinces) [15]. Co-existence of carbapenemase genes such as blaKPC, blaNDM, and blaVIM and mcr-1 in CRE isolated from humans, animals (chickens, ducks, and cats), and environmental samples has been reported [16–18]. Nevertheless, according to previous studies, the colistin resistance rate was not particularly high in CRKp (1.1%) and CREc (2.3%), and carriage of the mcr-1 gene among CRE remains rare in China [2,19].
To better understand the epidemiological trends and characteristics of MCR-1-producing clinical CRE strains collected before and after polymyxin was approved for use as an antimicrobial agent in clinical practices in China, we conducted extensive and systematic sampling in 24 provinces and municipalities in a span of 5 years (April 2014–April 2019). Findings in this work shall provide essential insight into development of effective strategies for worldwide control of mcr-1-carrying CRE and reduce the rate of untreatable hospital infections.
Materials and methods
Study design
To investigate the carriage of CRE, especially mcr-1-carrying CRE among hospital patients, we performed a cross-sectional multi-center study on samples collected from 1 January 2014 to 30 April 2019. Non-duplicated Enterobacteriaceae strains that exhibited carbapenem resistance phenotype (meropenem MIC ≥ 4 µg/mL) were collected from infection sites and clinical specimens of the patients including blood, urine, sputum, bile, hydrothorax, ascites, and various other specimens. All consecutive CRE isolates in the selected hospitals were stored and tested in the study period. A total of 1868 strains were collected from hospitals located in 24 provinces and municipalities in China including Anhui, Beijing, Fujian, Gansu, Guangdong, Guangxi, Guizhou, Hainan, Hebei, Henan, Hubei, Hunan, Jilin, Jiangxi, Liaoning, Jiangsu, Shandong, Shanxi, Shaanxi, Shanghai, Sichuan, Tianjin, Xinjiang, and Zhejiang (Table S1). These areas cover a population of 1.23 billion (∼90%) in China. One representative hospital, namely the largest general hospital in each location, was chosen for sample collection. Polymyxin has been applied in all these hospitals in 1 December 2017. All strains were subjected to species identification using the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany).
Gene screening and antimicrobial susceptibility testing
Carriage of carbapenem resistance genes (blaVIM, blaIMP, blaKPC, blaNDM, and blaoxa-48-like) and the colistin resistance gene mcr-1 to mcr-9 in all CRE isolates were screened by PCR, using methods described previously [14,20–22]. The genetic identity of the PCR products was validated using Sanger sequencing. All mcr-1-positive CRE strains were subjected to antimicrobial susceptibility testing against 15 commonly used antibiotics, using micro-broth dilution method. The minimum inhibitory concentrations (MICs) for carbapenems (imipenem, meropenem, ertapenem), cephems (cefepime, ceftazidime, cefotaxime, cefmetazole), fluoroquinolone (ciprofloxacin), β-lactam/β-lactamase combinations (piperacillin–tazobactam, cefoperazone–sulbactam (2:1), ceftazidime–avibactam), aminoglycoside (amikacin), and monobactam (aztreonam) were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [23]. Breakpoints for colistin and tigecycline were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria [24].
Whole-genome sequencing and bioinformatics analysis
All mcr-1-positive CRE isolates were subjected to whole-genome sequencing. Genomic DNA of each test isolate was extracted from overnight cultures using the PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). Genomic libraries were prepared with an insert of a size of ∼350 bp by the TruSeq DNA PCR-free Sample Preparation Kit (Illumina Inc., San Diego, CA, USA), following the manufacturer’s instructions and sequenced on the Illumina HiSeq 2500 platform (Annoroad Biotech Co.). Raw reads were trimmed with Trimmomatic to remove low-quality sequences and adaptors [25]. De novo assembly was conducted with SPAdes Genome Assembler v3.12.1 [26]. Assembled genome sequences were annotated with RAST [27]. Mobile antibiotic resistance genes were identified with ResFinder 2.1 [28]. Plasmid replicons were analysed using PlasmidFinder [29]. Insertion sequences (ISs) were identified using ISfinder [30]. Multilocus sequence typing and serotyping were conducted using MLST v2.11 and ECTyper v0.8.1, respectively [31,32]. Plasmid alignment was conducted using BRIG [33]. The harvest suite v.1.2 was used to remove recombination sequences and conduct the phylogenetic analysis using the assembled genome sequences as input and the genome sequence of strain SC-6 was used as a reference [34]. Phylogenetic tree was visualized and modified using iTOL v4 [35]. All draft genome sequences have been deposited in GenBank under BioProject accession number PRJNA558538.
Quantitative real-time PCR
Total RNA of mcr-1-positive strains was extracted using the RNeasy Mini kit (Qiagen, Hilden, Germany). Residual DNA was removed from the total RNA samples, and cDNA was synthesized with a PrimeScript RT Reagent Kit with gDNA eraser (TAKARA, Dalian, China) following the manufacturer’s instruction. Quantitative RT-PCR assay was performed using the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) kit (TAKARA) with primers published previously [36]. The 23S rRNA gene was used as the endogenous reference gene. Escherichia coli ATCC 25922 with a colistin MIC of 0.25 μg/mL was used as the reference strain. Relative expression level of the mcr-1 gene was obtained by the ΔΔCT analysis method.
Statistical analysis
The differences of mcr-1 carrying rate before and after December 2017 were assessed by Chi-Square Tests on IBM SPSS Statistics 20, and the selected specific chi-square test type depends on the expected count and the sample size of valid cases. Besides, the p values of <0.05 were considered to be statistically significant.
Results
Overview of the CRE strains
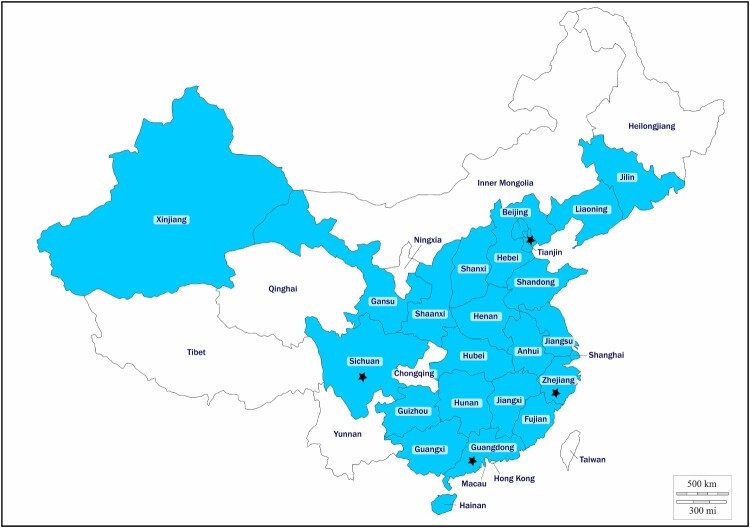
A total of 1868 non-duplicated CRE isolates were obtained from hospitals in 24 provinces and municipalities in China, including 1134 K. pneumoniae isolates, 376 E. coli isolates, 184 Enterobacter cloacae isolates, 41 Serratia marcescens isolates, 45 Enterobacter aerogenes isolates, 31 K. oxytoca isolates, 29 Citrobacter freundii isolates, and 28 other isolates (C. koseri, C. braakii, Raoultella ornithinolytica, Providencia rettgeri, and Proteus mirabilis) (Table 1). The number of strains isolated before and after 21 February 2017 were 1215 and 653, respectively. PCR assay indicated that 1755 (96.18%) isolates carried the carbapenemase genes blaKPC and blaNDM, and that 14 isolates (0.75%) were positive for mcr-1. No strain positive for other mcr variants was detected. The 14 mcr-1 carrying isolates, all identified as E. coli and termed MCR-CREC hereafter, were isolated from sputum, blood, drainage, secretion, pus, and throat swab specimens (Table 2). The blaNDM gene, but not other carbapenem resistance genes, were identified in all the MCR-CREC isolates. Among these 14 blaNDM genes, 2 (14.29%) were blaNDM-4, 11 (78.57%) were blaNDM-5, and 1 (7.14%) was blaNDM-9. The MCR-CREC isolates were recovered from four provinces or regions, namely Zhejiang (n = 9, 64.29%), Tianjin (n = 2, 14.29%), Guangdong (n = 2, 14.29%), and Sichuan (n = 1, 7.14%) (Figure 1, Table 2). The number and proportion of MCR-CREC isolates among the test strains were found to increase from 5 (0.41%) to 9 (1.38%) after introduction of polymyxin into clinical practice, such increase was statistically significant (P < 0.05). MCR-CREC were detectable in all the four provinces mentioned above before 1 December 2017; however, eight of the nine (88.9%) MCR-CREC strains isolated thereafter were collected from Zhejiang province, indicating that a rather localized dissemination of MCR-CREC strains had occurred, presumably due to antibiotic selection.
Table 1. Distribution of mcr-1 in CREs during 2014–2019.
| Strain species | No. of strains (1 April 2014–20 February 2017) | No. of strains (21 February 2017–30 April 2019) | P-value* | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRE | blaKPC positive | blaNDM positive | CPE | mcr-1 |
mcr-1 carrying rate (%) |
CRE | blaKPC positive | blaNDM positive | CPE | mcr-1 |
mcr-1 carrying rate (%) |
||
| K. pneumoniae | 740 | 554 | 121 | 705 | 0 | 0 | 394 | 309 | 32 | 384 | 0 | 0 | – |
| E. coli | 235 | 85 | 132 | 221 | 5 | 2.13% | 141 | 62 | 73 | 137 | 9 | 6.38% | 0.035 |
| Enterobacter cloacae | 132 | 19 | 81 | 105 | 0 | 0 | 52 | 9 | 39 | 52 | 0 | 0 | – |
| Serratia marcescens | 21 | 6 | 14 | 21 | 0 | 0 | 20 | 7 | 0 | 7 | 0 | 0 | – |
| Enterobacter aerogenes | 26 | 6 | 16 | 26 | 0 | 0 | 19 | 15 | 1 | 16 | 0 | 0 | – |
| Citrobacter freundii | 21 | 2 | 13 | 21 | 0 | 0 | 8 | 3 | 5 | 8 | 0 | 0 | – |
| K. oxytoca | 24 | 7 | 10 | 24 | 0 | 0 | 7 | 2 | 3 | 5 | 0 | 0 | – |
| Other species | 16 | 7 | 7 | 12 | 0 | 0 | 12 | 6 | 5 | 11 | 0 | 0 | – |
| Total | 1215 | 686 | 394 | 1135 | 5 | 0.41% | 653 | 413 | 158 | 620 | 9 | 1.38% | 0.042 |
CRE, carbapenem-resistant Enterobacteriaceae; CPE, carbapenem-producing Enterobacteriacease.
*Differences between groups were assessed by Pearson chi-square and continuity correction. P-value in bold indicates statistical significance.
Table 2. Strain source, susceptibility to commonly used antibiotics and mcr-1 expression levels of mcr-1-positive isolates.
| Bacterial | Year | provinces | specimen | MLST | Serotype | mcr-1- plasmid | blaNDM- plasmid | MIC (mg/L) | ΔΔCt expression** (average ± StdDev) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ETP | CMZ | CAZ | CTX | TZP | SCF | CAV | FEP | PE | TGC | CIP | AK | ATM | |||||||||
| 1 April 2014–20 February | |||||||||||||||||||||||
| TJ-2 | 2015 | Tianjin | sputum | 167 | O89:H9 | IncX4 | Na* | 32 | 64 | 128 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | ≤0.5 | ≤0.25 | >32 | >128 | 32 | 2.279 ± 0.547 |
| GD133 | 2015 | Guangdong | blood | 10 | O89:H10 | IncHI2 | IncX3 | 16 | 16 | 32 | 16 | >128 | >128 | 256/4 | 256/4 | >64/4 | 64 | 1 | ≤0.25 | 32 | ≤4 | 16 | 227.873 ± 41.474 |
| GD119 | 2015 | Guangdong | sputum | 10 | O89:H10 | IncHI2 | IncX3 | 16 | 64 | 64 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | ≤0.5 | ≤0.25 | >32 | ≤4 | >128 | 289.955 ± 55.608 |
| SC-6 | 2015 | Sichuan | sputum | 167 | O89:H9 | IncX4 | IncX3 | 32 | 128 | 128 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | 2 | ≤0.25 | 32 | ≤4 | 16 | 107.323 ± 17.276 |
| C34 | 2015 | Zhejiang | Secretion | 5229 | O177:H51 | IncX4 | IncX3 | 32 | 128 | 128 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | ≤0.5 | ≤0.25 | >32 | >128 | >128 | Na. * |
| 21 February 2017–30 April 2019 | |||||||||||||||||||||||
| TJ-71 | 2017 | Tianjin | Drainage | 617 | O89:H9 | IncI2 | IncFII | 8 | 32 | 64 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | 2 | ≤0.25 | 32 | >128 | >128 | 107.539 ± 19.591 |
| 363 | 2017 | Zhejiang | sputum | 4380 | O116:H9 | IncHI2A/HI2 | Na* | 16 | 32 | 32 | 64 | >128 | >128 | 256/4 | >256/4 | >64/4 | 64 | 2 | ≤0.25 | 32 | ≤4 | ≤4 | 133.119 ± 15.133 |
| 404 | 2017 | Zhejiang | sputum | 23 | O78:H9 | IncHI2 | IncX3 | 8 | 16 | 32 | 16 | >128 | >128 | >256/4 | 256/4 | >64/4 | 64 | 2 | ≤0.25 | ≤1 | ≤4 | ≤4 | 448.233 ± 116.063 |
| 34 | 2017 | Zhejiang | sputum | 10 | O60:H12 | IncX4 | IncX4 | 8 | 16 | 32 | 16 | >128 | >128 | 128/4 | 256/4 | >64/4 | 64 | 2 | ≤0.25 | ≤1 | ≤4 | ≤4 | 522.624 ± 84.262 |
| 76 | 2017 | Zhejiang | Pus | 2179 | O9:H9 | IncHI2 | IncX3 | 16 | 32 | 32 | 8 | >128 | >128 | 128/4 | 256/4 | >64/4 | 32 | 2 | ≤0.25 | 16 | ≤4 | ≤4 | 246.972 ± 39.069 |
| 182 | 2017 | Zhejiang | Throat swab | 189 | O37:H21 | IncY | IncX3 | 8 | 16 | 32 | 32 | >128 | >128 | 128/4 | 256/4 | >64/4 | 64 | 2 | ≤0.25 | 16 | ≤4 | ≤4 | 107.892 ± 21.309 |
| 362–1 | 2017 | Zhejiang | blood | 4380 | O116:H9 | IncHI2A/HI2 | Na* | 8 | 8 | 16 | 128 | >128 | >128 | 128/4 | 128/4 | >64/4 | 16 | 2 | ≤0.25 | ≤1 | ≤4 | 32 | 189.66 ± 43.697 |
| shaw79 | 2019 | Zhejiang | sputum | 617 | O89:H9 | IncI2 | IncFII | 8 | 16 | 64 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | 2 | ≤0.25 | >32 | ≤4 | ≤4 | 194.778 ± 43.785 |
| sz16 | 2019 | Zhejiang | blood | 297 | Ounknown:H9 | IncI2 | IncFII | 8 | 32 | 32 | >128 | >128 | >128 | >256/4 | >256/4 | >64/4 | >64 | 2 | ≤0.25 | 8 | ≤4 | >128 | 190.368 ± 35.552 |
IMP, imipenem; MEM, meropenem; ETP, ertapenem; CMZ, cefmetazole; CAZ, ceftazidime; CTX, cefotaxime; TZP, piperacillin/tazobactam; SCF, cefoperazone/sulbactam; CAV, ceftazidime/avibactam; FEP, cefepime; PE, polymyxin E; TGC, tigecycline; CIP, ciprofloxacin; AK, amikacin; ATM, aztreonam.
*Na, not applicable; **ΔΔCt here was used to describe the relative mcr-1 gene expression levels in the strains.
Figure 1.
Distribution of mcr-1 in carbapenem-resistant Enterobacteriaceae strains in China. mcr-1-producing isolates were denoted with stars. The provinces included in this study are shaded.
Antimicrobial susceptibility of MCR-CREC and expression level of mcr-1
The 14 MCR-CREC isolates were all resistant to carbapenems but were susceptible to tigecycline. The proportion of MCR-CREC isolates that were resistant to colistin, ciprofloxacin, amikacin, and aztreonam were 71.4%, 78.6%, 21.4%, and 57.1%, respectively (Table 2). Among the five MCR-CREC strains isolated before 1 December 2017, only one (20%) was resistant to colistin (MIC = 2 mg/mL), one (20%) exhibited intermediate resistance to colistin (MIC = 1 mg/mL), and the other three (60%) remained susceptible to colistin (MIC ≤0.5 mg/mL). However, all nine MCR-CREC strains isolated after 1 December 2017 were resistant to colistin, exhibiting an MIC of 2 mg/mL (Table 2, Fig S1) indicating that colistin MICs for mcr-1-positive CREC increased after clinical use of colistin.
To test whether the clinical application of colistin selected the expression of mcr-1, relative expression levels of mcr-1 was measured by qRT-PCR assays (Table 2, Fig S2). Strain C34 was excluded in the qPCR experiment due to the similar resistance profiles of strains C34 and TJ-2. Among the 13 strains tested, TJ-2 isolated in 2015 exhibited the lowest mcr-1 expression level. The expression levels of mcr-1 gene in other strains isolated before 1 December 2017 were 47, 100, and 127 folds of that of TJ-2. That of strains isolated after 1 December 2017 were 47 (2 strains), 58, 83 (2 strains), 85, 108, 197, and 224 folds of strain TJ-2. With these data, we could not determine whether the use of colistin select the expression of mcr-1. Despite strains isolated after 1 December 2017 exhibited identical susceptibility phenotypes to colistin (MIC = 2 mg/L), the expression levels of mcr-1 varied dramatically among strains, indicating the expression of mcr-1 was potentially under regulation of other genetic components, which warrants further investigation.
Genetic background of MCR-CREC
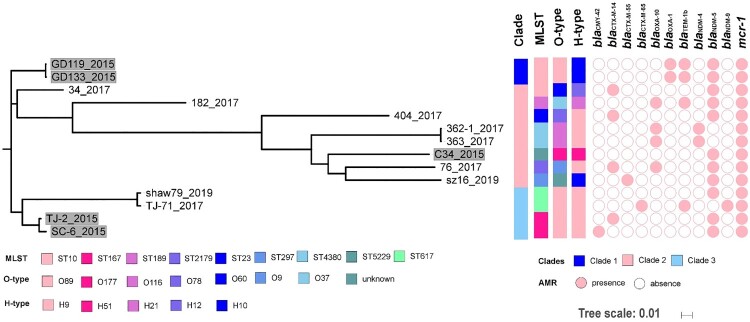
The 14 MCR-CREC isolates were allocated to nine ST types and different serotypes (eight O types and five H types, Table 2, Figure 2). None of these ST types was dominant, but ST167 (n = 2), ST617 (n = 2), ST4380 (n = 2), and ST10 (n = 2) were more prevalent than the other sequence types. A total of 60,534 SNPs (minimum: 34; maximum: 29698) were identified among the core genomes of these isolates, on the basis of which the isolates could be allocated into least three major lineages (Fig S3). The first clade includes two ST10 E. coli isolates (GD119, GD133) both isolated before 1 December 2017. The second clade represents a major branch of mcr-1-bearing carbapenem-resistant E. coli, which comprised eight isolates (34, 182, 404, 362-1, 363, C34, 76, sz16) of seven sequence types, with seven out of the eight isolates being isolated after 1 December 2017. The third clade includes four isolates, two isolated before (n = 2, ST167) and two after (n = 2, ST617) December 2017. The data indicated that MCR-CREC strains were genetically diverse and covered all three genetic clades before the use of colistin in clinical setting with only one strain belonging to clade II, yet most of the strains after the use of colistin in clinical setting were selected in clade II. When looked at the serotypes, four strains of O89 belonging to clade I and III were isolated before use of colistin, while more diverse O types in clade II were found in mcr-1-positive CREC after the use of colistin.
Figure 2.
Mid-point rooted phylogeny of mcr-1-bearing E. coli strains. (A) Strains isolated before 1 December 2017 were labelled with grey backgrounds. MLST, O and H serotypes, the presence of antimicrobial resistance genes (AMR) encoding ESBLs, carbapenemases, and MCR-1 were plotted. Year of isolation for each strain was denoted after the dash line.
Multiple antimicrobial resistance genes, including ESBLs (blaCMY, blaCTX, blaTEM, and blaOXA), carbapenemase gene blaNDM and the mcr-1 gene, were detectable among the test isolates (Figure 2).
To further investigate the role of MCR-CREC strains in dissemination of the blaNDM and mcr-1 genes, we analysed the draft whole-genome sequence of the 14 MCR-CREC strains and found that the mcr-1 gene was located in contigs ranging from 2 to 109 kb. The mcr-1-bearing contigs in MCR-CREC aligned well to different plasmids, including the 97 kb IncY pHYEC7-MCR1 plasmid (accession: KX518745, n = 1), the 169 kb IncHI2A/HI2 pHNSHP47 plasmid (accession: MF774186, n = 2), the 251 kb IncHI2 pHNSHP45-2 plasmid (accession: KU341381, n = 4), the 33 kb IncX4 pESTMCR plasmid (accession: KU743383, n = 3), and the 63 kb IncI2 pOM97-mcr plasmid (accession: KY693674, n = 3) (Fig S4). IncX4 and IncHI2 types of mcr-1-bearing plasmids were detected in these CREC isolates, while three more plasmids including IncI2, IncHI2A/HI2, and IncY were detected in isolates after the use of colistin (Table 2). BLAST of the blaNDM genes in the draft genomes of these strains indicated that the gene was located on contigs ranging from 2 to 43 kb. Genomic contents of the 14 strains aligned well to the 46 kb IncX3 plasmid pNDM-EC36 (accession: MG591703, n = 7) and the 116 Kb IncFII plasmid pEC1107-NDM-116 K (accession: MG601057, n = 3), separately (Fig S5). Due to poor assembly quality, contigs carrying blaNDM in three isolates (TJ-2, 221, 362-1, and 363) were too short to resolve the picture of the plasmid harbouring blaNDM. Besides, co-existence of mcr-1 and blaNDM in one contig was detected in one isolate, 34, which was isolated before December 2017. The contig bearing both genes aligned well to the 48 kb IncX4 plasmid pCQ02-121 (accession: KU647721, Fig S6). For isolates carrying known types of blaNDM-bearing plasmids, all isolates before the use of colistin carried incX3 types of plasmids, while IncFII and IncX4 type of plasmid were found in addition to IncX3 type in CREC isolates after the use of colistin (Table 2).
Discussion
Since the discovery of the plasmid-mediated polymyxin resistance gene mcr-1 at the end of 2015, mobile colistin resistance determinants have attracted worldwide attention [14]. To date, nine different mcr variants, namely mcr-1 to mcr-9, have been documented [22]. These mcr genes have been disseminated worldwide and were reported to coexist with many different resistance genes, especially carbapenem resistance genes (blaNDM, blaKPC, etc.), resulting in the emergence of veritable “superbugs” which pose severe threats to human health [37–41]. To our knowledge, this study was the first nationwide surveillance report comparing the carriage of mcr-1 among CRE before and after use of polymyxin in clinical settings.
The prevalence of colistin resistance was relatively low in Enterobacteriaceae strains in China and many other countries [42,43]. In this study, although the mcr-1 carriage rate was not particularly high among CRE (0.75%), our data show that the percentage of CRE isolates that carry mcr-1 increased dramatically from 0.41% to 1.38% after polymyxin was adopted for clinical use, indicating the use of colistin prompted the emergence and spread of MCR-CRE in China. Most MCR-CREC strains (n = 9) were isolated from Zhejiang province, probably due to the relatively high antimicrobial resistance rates in Zhejiang and a large proportion of strains (28%, 524/1868) was collected from Zhejiang province. Alarmingly, we found the resistance level of MCR-CREC isolates to colistin increased after the introduction of polymyxin into clinical use with the MICs to colistin increasing from <2 mg/L in 80% strains to 2 mg/L in 100% strains. The quantification of the mcr-1 gene was tricky due to strain differences, thus we could not conclude whether the use of colistin have selected the expression of mcr-1 in the CREC strains. But the worrying scenario of that mcr-1 as well as the colistin resistance level increased within a short period of time (2014–2019) is a reminder that we need to take urgent actions.
The blaKPC-2 and blaNDM was responsible for phenotypic resistance in 58% and 32% of the CRE strains in China, respectively [2]. Co-existence of mcr genes and different carbapenem resistance genes such as blaKPC-3, blaNDM-5, and blaNDM-16 have been sporadically reported in Enterobacteriaceae isolated from patients from different continents, among which E. coli was the most reported hosts [44–46]. Consistently, all mcr-1-positive CRE strains in this study belonged to E. coli, indicating E. coli was the major reservoir of such resistance genes. A previous study indicated acquisition of mcr-1 or blaNDM-5 plasmid did not lead to considerable fitness costs, highlighting the potential for dissemination of mcr-1 and blaNDM-5 in Enterobacteriaceae [47]. In this study, mcr-1 and blaNDM-5 are the most dominant (78.6%, 11/14) combination among MCR-CREC strains. Escherichia coli isolates carrying both mcr-1 and blaNDM-5 also have disseminated among healthy people in various regions of China [48]. The pattern of transmission of resistance genes between patients and healthy individuals should also be carefully monitored. More diverse blaNDM variants including blaNDM-4 and blaNDM-9 emerged in the clinical settings after the use of polymyxin, suggesting an alarming evolutionary trend of MCR-CREC strains.
Several different E. coli ST isolates such as ST131, ST405, ST167, ST206, ST648, ST10, ST617, and ST156 have been reported to carry both blaNDM variants and mcr genes [47,49,50]. In this study, three and seven E. coli ST types were detected before and after the use of colistin, respectively. The serotypes of the MCR-CREC strains were more diverse after use of colistin. These findings suggested the application of colistin in clinical settings expanded the host range of E. coli strains carrying both blaNDM variants and mcr-1 genes. blaNDM variants and mcr determinants are predominantly encoded by mobile genetic elements such as conjugative plasmids, transposons, and integrons, which are readily transferable [51]. The major plasmid types carrying mcr-1 published to date included IncX4, IncI2, IncHI2, IncY [52,53]. Diverse plasmid types carrying blaNDM variants have been reported, including IncX3, IncFII, IncH, and IncA/C [54–57]. The type of plasmids carrying blaNDM variants and mcr-1 both became more diverse under colistin treatment in clinical settings, indicating that colistin treatment prompted the transmission of diverse resistance plasmids among E. coli isolates. Besides, a strain (E. coli 34) with IncX4 plasmid carrying both blaNDM-5 and mcr-1 genes was isolated after December 2017. Co-transfer of blaNDM and mcr-1 by a single plasmid has been rare since the discovery of mcr-1, with only one case reporting a plasmid (pCQ02-121) similar to the one in this study generated from genetic recombination between the IncX3 blaNDM-5-carrying and the IncX4 mcr-1-carrying plasmid [41]. pCQ02-121 was from an ST156 E. coli strain isolated from a pet cat in China in 2015, and in this study, E. coli 34 was an ST10 strain isolated from a human carrier, suggesting the transmission potential of pCQ02-121-like plasmids within different host strains and among different carrier organisms such as humans and companion animals [41]. Heightened efforts are needed to control the dissemination of pCQ02-121-like plasmids. The potential of E. coli isolates to acquire plasmids and mobile elements carrying blaNDM and mcr-1 genes rendered them to become “superbugs” resistance to last-line antibiotics which posed a grave threat to human health.
Conclusions
This is the first comprehensive study which investigated the carriage of mcr-1 by CRE among individuals in the clinical settings. CRE strains positive for mcr-1 were all E. coli with diverse sequence types and serotypes. The official issue of polymyxin in hospitals in February 2017 lead to a significant increase in MCR-CREC isolates and elevated resistance profiles to polymyxins. NDM was the major carbapenemase carried by MCR-CREC, with NDM-5 being the most dominant type. Genomic analysis indicated the mcr-1 and blaNDM genes were carried by different plasmids which are highly transmissible. Due to the high prevalence of CRE and increasing carriage of MCR-CREC isolates among patients in the clinical settings, urgent actions should be taken to prevent the dissemination of such pathogens in high-risk patients.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China (81772250, 81861138052) and Collaborative Research Fund of Hong Kong Research Grant Council (C5026-16G).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Logan LK, Weinstein RA.. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang R, Liu L, Zhou H, et al. . Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Wang Q, Yin Y, et al. . Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2):e01882–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu F-P, Guo Y, Zhu DM, et al. . Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22:S9–S14. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Dortet L, Poirel L.. Carbapenem resistance in Enterobacteriaceae: here is the storm!. Trends Mol Med. 2012;18(5):263–272. [DOI] [PubMed] [Google Scholar]
- 6.Boutal H, Vogel A, Bernabeu S, et al. . A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP-and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2018;73(4):909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Chan EW-c, Zhou H, et al. . Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect Dis. 2017;17(3):256–257. [DOI] [PubMed] [Google Scholar]
- 8.van Duin D, Bonomo RA.. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis. 2016;63(2):234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He T, Wang R, Liu D, et al. . Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–1456. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization . Critically important antimicrobials for human medicine, 3rd revision. Geneva: WHO; 2012. [Google Scholar]
- 11.Giamarellou H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 2016;48(6):614–621. [DOI] [PubMed] [Google Scholar]
- 12.Ah Y-M, Kim A-J, Lee J-Y.. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2014;44(1):8–15. [DOI] [PubMed] [Google Scholar]
- 13.Walsh TR, Wu Y.. China bans colistin as a feed additive for animals. Lancet Infect Dis. 2016;16(10):1102–1103. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y-Y, Wang Y, Walsh TR, et al. . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Zhou H, Xu J, et al. . Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol. 2018;3(9):1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhang R, Li J, et al. . Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2(4):16260. [DOI] [PubMed] [Google Scholar]
- 17.Sun P, Bi Z, Nilsson M, et al. . Occurrence of blaKPC-2, blaCTX-M, and mcr-1 in Enterobacteriaceae from well water in rural China. Antimicrob Agents Chemother. 2017;61(4):e02569–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bocanegra-Ibarias P, Garza-González E, Morfín-Otero R, et al. . Molecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in Mexico. PLoS ONE. 2017;12(6):e0179651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castanheira M, Griffin MA, Deshpande LM, et al. . Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY antimicrobial surveillance program during 2014-2015. Antimicrob Agents Chemother. 2016: AAC. 01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallenne C, Da Costa A, Decré D, et al. . Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. [DOI] [PubMed] [Google Scholar]
- 21.Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. . Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance. 2018;23(6):17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieffer N, Royer G, Decousser JW, et al. . mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother. 2019: AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and L.S. Institute. Performance standards for antimicrobial testing: twenty-sixth informational supplement M100-S26. Wayne, PA: CLSI; 2016.
- 24.Testing E.C.o.A.S: breakpoint tables for interpretation of MICs and zone diameters. 2016, version.
- 25.Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overbeek R, Olson R, Pusch GD, et al. . The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2013;42(D1):D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zankari E, Hasman H, Cosentino S, et al. . Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carattoli A, Zankari E, Garcìa-Fernandez A, et al. . Plasmidfinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother. 2014: AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siguier P, Pérochon J, Lestrade L, et al. . ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(suppl_1):D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Scan contig files against PubMLST typing schemes. 2016. Available from: https://github.com/tseemann/mlst.
- 32.Github. ECTyper. Available from: https://github.com/phac-nml/ecoli_serotyping.
- 33.Alikhan N-F, Li Y, Lee W, et al. . BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treangen TJ, Ondov BD, Koren S, et al. . The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I, Bork P.. Interactive tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nezhadi J, Narenji H, Soroush Barhaghi MH, et al. . Peptide nucleic acid-mediated re-sensitization of colistin resistance Escherichia coli KP81 harboring mcr-1 plasmid. Microb Pathog. 2019;135:103646. [DOI] [PubMed] [Google Scholar]
- 37.Aires CAM, da Conceição-Neto OC, e Oliveira TRT, et al. . Emergence of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae sequence type 392 in Brazil. Antimicrob Agents Chemother. 2017;61(7):e00317–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalmolin TV, Castro L, Mayer FQ, et al. . Co-occurrence of mcr-1 and bla KPC-2 in a clinical isolate of Escherichia coli in Brazil. J Antimicrob Chemother. 2017;72(8):2404–2406. [DOI] [PubMed] [Google Scholar]
- 39.Delgado-Blas JF, Ovejero CM, Abadia-Patiño L, et al. . Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60(10):6356–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong L-H, Lei CW, Ma SZ, et al. . Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob Agents Chemother. 2017;61(3):e02167–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Yang R-S, Zhang Q, et al. . Co-transfer of bla NDM-5 and mcr-1 by an IncX3–X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1(12):16176. [DOI] [PubMed] [Google Scholar]
- 42.Quan J, Li X, Chen Y, et al. . Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. [DOI] [PubMed] [Google Scholar]
- 43.Falagas ME, Rafailidis PI, Matthaiou DK.. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist Updates. 2010;13(4–5):132–138. [DOI] [PubMed] [Google Scholar]
- 44.Tacao M, Tavares RdS, Teixeira P, et al. . mcr-1 and blaKPC-3 in Escherichia coli sequence type 744 after meropenem and colistin therapy, Portugal. Emerging Infect Dis.. 2017;23(8):1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mediavilla JR, Patrawalla A, Chen L, et al. . Colistin-and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4):e01191–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Mu X, Zhang P, et al. . Detection and characterization of a clinical Escherichia coli sT3204 strain coproducing nDM-16 and McR-1. Infect Drug Resist. 2018;11:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Liao K, Gao H, et al. . Decreased fitness and virulence in ST10 Escherichia coli harboring blaNDM-5 and mcr-1 against a ST4981 strain with blaNDM-5. Front Cell Infect Microbiol. 2017;7:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, Hu Y, Sun Q, et al. . Emerging carriage of NDM-5 and MCR-1 in Escherichia coli from healthy people in multiple regions in China: a cross sectional observational study. EClinicalMedicine. 2018;6:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng B, Yu X, Xu H, et al. . Complete genome sequencing and genomic characterization of two Escherichia coli strains co-producing MCR-1 and NDM-1 from bloodstream infection. Sci Rep. 2017;7(1):17885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen LP, Pinto NA, Vu TN, et al. . Resistome profiles, plasmid typing, and whole-genome phylogenetic tree analyses of BlaNDM-9 and Mcr-1 Co-harboring Escherichia coli ST617 from a patient without a history of farm exposure in Korea. Pathogens. 2019;8(4):212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong N, Zhang R, Liu L, et al. . Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microb Genomics. 2018;4(2):e000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Feng Y, Liu F, et al. . A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob Agents Chemother. 2017;61(3):e02035–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li R, Xie M, Zhang J, et al. . Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2016;72(2):393–401. [DOI] [PubMed] [Google Scholar]
- 54.Villa L, Poirel L, Nordmann P, et al. . Complete sequencing of an IncH plasmid carrying the bla NDM-1, bla CTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67(7):1645–1650. [DOI] [PubMed] [Google Scholar]
- 55.Carattoli A, Villa L, Poirel L, et al. . Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother. 2012;56(2):783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonnin RA, Poirel L, Carattoli A, et al. . Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE. 2012;7(4):e34752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho P-L, Li Z, Lo WU, et al. . Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect. 2012;1(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.