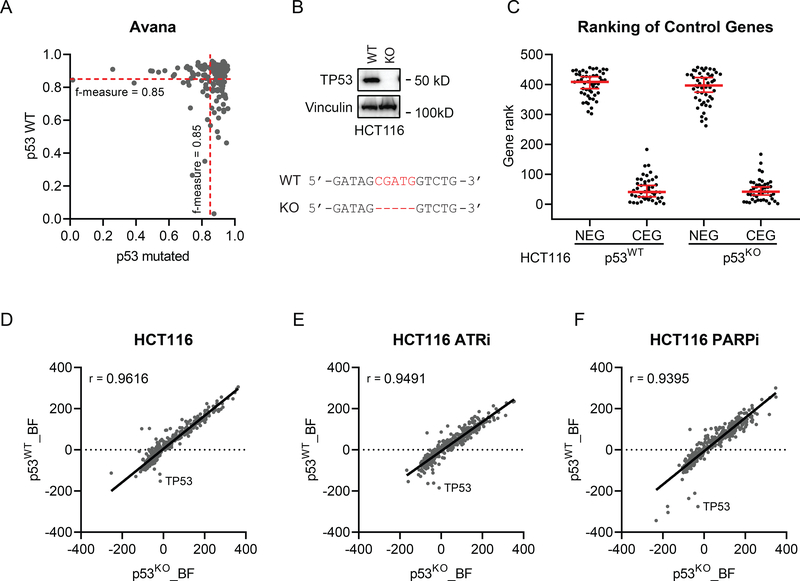
Figure 6. Intact TP53 does not influence the performance of CRISPR screens.
(A) Similar CRISPR screen performance was noted in TP53 wild-type (WT) and TP53-mutated cancer cell lines. One hundred eighty-three Avana screens performed in TP53 WT cell lines were compared with randomly sampled same number of screens performed TP53 mutant cell lines. (B) Western blotting verified TP53 knockout (KO) in HCT116 cells. Vinculin was used as the loading control. Sanger sequencing showed that 5 nt (CGATG) of TP53 coding sequence was deleted in HCT116 KO cells. (C) The gene ranking of core-essential genes (CEG) and nonessential genes (NEG) in HCT116 WT and TP53 KO cells based on a log Bayes factor (BF) calculated using the BAGEL approach. Red line indicates median rank and 95% confidence interval. (D-F) Spearman correlation coefficient of BF distributions of DNA damage response genes in HCT116 WT and KO cells in mock treated (D), ATR inhibitor (ATRi)-treated (E), and PARP inhibitor (PARPi)-treated groups (F).