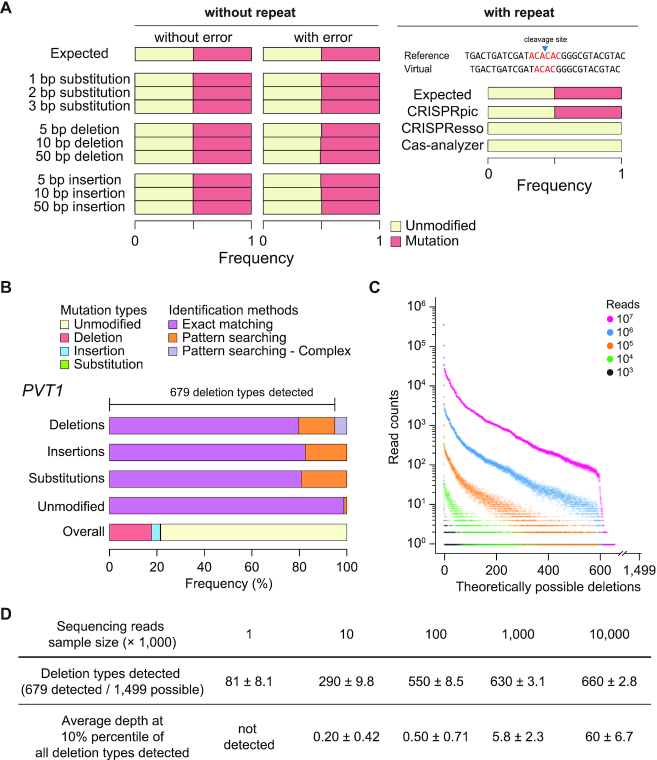
Figure 2.
Pilot analysis using CRISPRpic for indel identification. (A) Simulation analysis using CRISPRpic. Simulation results using virtual sequencing reads with or without repetitive sequences around the cleavage site. AC repeats are shown in red. (B) Mutation analysis of sequencing reads from the PVT1 locus targeted by SpCas9. Bar plots showing proportions of mutation callings by exact matching or pattern searching. (C) Distribution of all theoretically possible deletions at the PVT1 locus by sample size of sequencing reads (n = 10, technical replicates). (D) Summary for coverage and sequencing depth of detection of deletions.