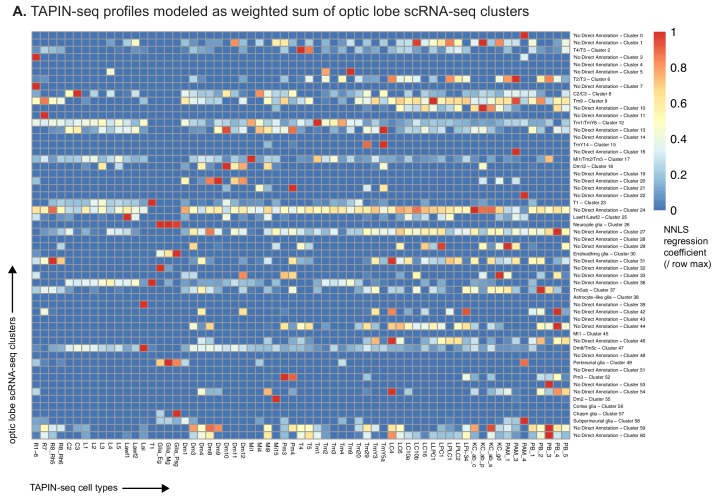
Figure 5. TAPIN-seq complements single cell RNA-seq profiling.
(A, B) We evaluated whether single cell RNA-seq of the optic lobe (A) (Konstantinides et al., 2018) and brain (Davie et al., 2018) proportionally represents cell types found in the optic lobe. By comparing the single cell cluster sizes to the true abundance of each cell type (estimated as described in the Materials and methods) we found that the scRNA-seq map can both under- and over-estimate the abundance of each cell type (assuming accurate cell type labels), or that the cell type is incorrectly assigned (i.e. contains different or additional cell types). To estimate the true cell count, we made use of known anatomy (for example, several cell types are known to be present exactly once in each of the ~2×750 medulla columns per brain) or relied on published counts. In addition, we performed some new counts. (See Materials and methods for details.) Observed/expected ratio = ((size of cluster labeled as cell type X/size of cluster labeled as T1) / (true abundance of cell type X/true abundance of T1)). (C) We used non-negative least squares regression to model each TAPIN-seq profile as a linear weighted sum of single cell clusters in the whole brain scRNA-seq map. The heatmap represents the regression coefficients of each single cell cluster (rows) contributing to the TAPIN-seq profile of each cell type, normalized within rows. (D) We evaluated expression of genes that mark selected single cell clusters (Davie et al., 2018) in our TAPIN-seq profiles of visual system neurons. (see Figure 5—figure supplement 2 for the complete heatmap).