Fig. 4. Mutations demonstrate nonequivalent PU.1 dimers with and without DNA.
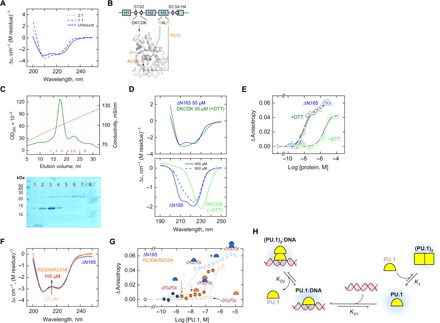
(A) Far-UV CD spectra of the DNA-bound ΔN165 upon subtracting the spectrum of the cognate DNA acquired under identical conditions (75 μM and 0.15 M Na+). (B) Residues involved in the DKCDK mutant and in the binding-deficient mutant (R230A/R233A). The structure is homology-modeled against the cocrystal 1PUE. (C) Purification of the DKCDK mutant by ion exchange chromatography under nonreducing conditions. Lysate was loaded at 0.5 M NaCl and extensively washed before elution over a linear gradient to 2 M NaCl. SDS-PAGE of purified fractions is shown. Fractions containing primarily monomer (e.g., 1 and 2) or dimers (e.g., 5 onwards) were concentrated and dialyzed separately into buffer containing 0.15 M NaCl with or without 5 mM DTT, respectively. (D) CD spectra of the DKCDK monomer (top) and dimer (bottom) under various conditions with wild-type ΔN165 as reference. The spectrum for the DKCDK monomer was less well resolved due to the presence of DTT, which contributed to the total absorption of the sample at 50 μM protein. See text for details. (E) Fluorescence anisotropy measurements of cognate DNA binding by monomeric and dimeric DKCDK with wild-type ΔN165 as reference. (F) CD spectrum of 25 to 100 μM of the R230A/R233A mutant, with ΔN165 at 25 μM as reference. (G) DNA loading by the R230A/R233A mutant in the presence of wild-type ΔN165 (solid symbols). Concentrations of the mutant and wild-type protein that individually failed to bind DNA collaborated to bind DNA as a heterocomplex. (H) Proposed model for the formation of two nonequivalent PU.1 dimers: an asymmetric one in the 2:1 DNA complex and a symmetric one without DNA.
