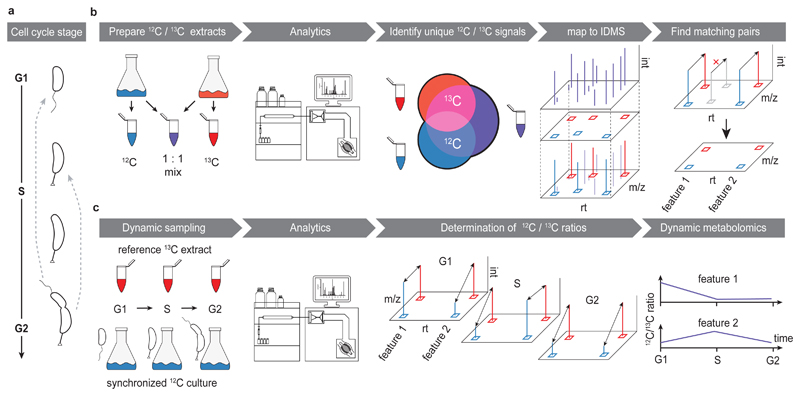
Figure 1. Untargeted dynamic metabolomics to monitor cell cycle progression.
(a) Schematics of the C. crescentus cell cycle and differentiation. Cell cycle progression is linked to clearly distinguishable changes in morphology. A motile swarmer cell (G1) sheds its flagellum, and develops into a stalked, proliferative cell that undergoes replication (S), and ultimately divides (G2) into new swarmer cell, while itself remaining a stalked cell that re-initiates S phase. (b) Building a non-targeted metabolite library. Production of naturally 12C - (blue) and highly uniformly (u) 13C -isotope enriched (red) C. crescentus extracts by growth on respectively labelled carbon sources to obtain a low- and high-molecular-weight metabolome. Accessible peaks from both extracts, as well as a mix thereof (12C/13C mix, purple), were subsequently detected using LC-HRMS that distinguishes mass shifts associated with isotope incorporation. Shared peaks between 12C and 13C samples were discarded as spectral noise. Unique peaks with a clear isotopic identity (u-12C, u-13C) were mapped to the peakmap of the mixed sample. Lastly, this peakmap was filtered by matching co-eluting peaks that were separated by a m/z shift explained by carbon labelling. This approach selected features of biological origin, and provided the extraction windows for the subsequent, isotope-dilution based metabolomics approach. (c) Dynamic metabolomics of the C. crescentus cell cycle. Synchronized cells growing on 12C glucose were followed throughout one cell cycle. For each time point, a 12C aliquot normalized to biomass was sampled and spiked with a constant amount of reference 13C enriched C. crescentus extract. Changes in relative pool sizes over time were then determined by calculating the peak area ratios of the u-12C relative to corresponding u-13C peak areas.