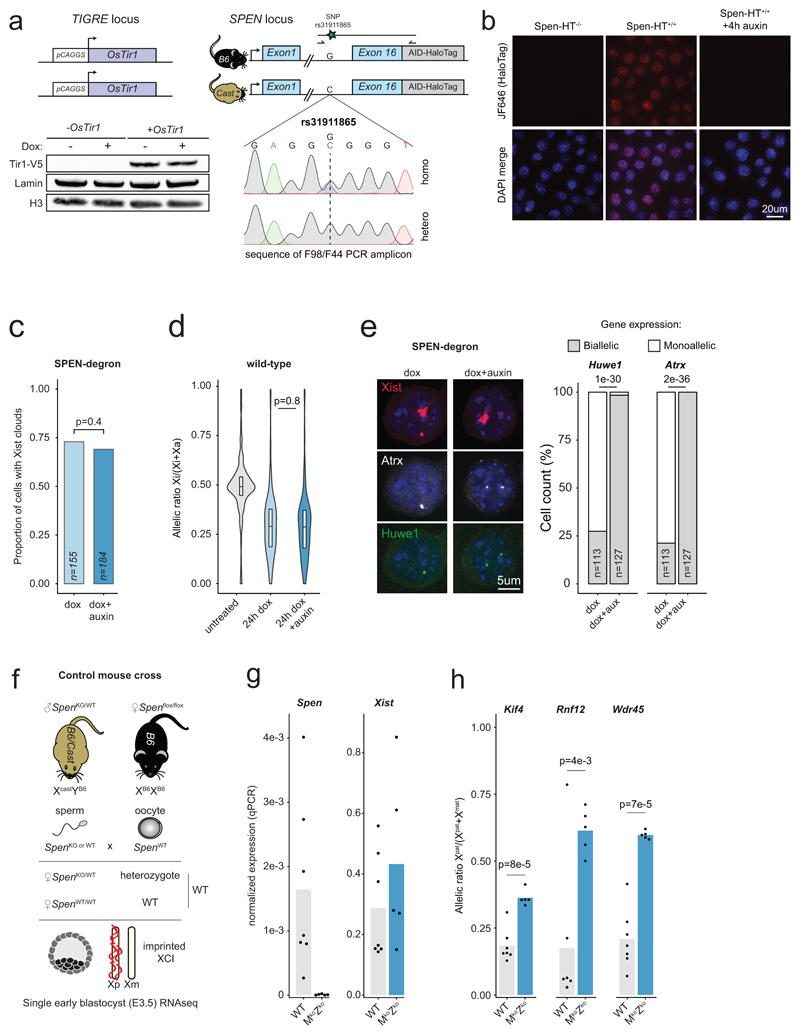
Extended Data Figure 1. SPEN mediates gene silencing across the entire X chromosome in vitro and in vivo.
a, Schematic representation of the SPEN-degron genotype with AID-HaloTag insertions in frame with the C-terminus of endogenous SPEN. Targeted homozygous insertion of V5-tagged OsTir1 at the TIGRE locus (top left) results in its constitutive protein expression as assessed by western blot (bottom left). Sanger sequencing results for a PCR amplicon specific to AID-HaloTag insertions and covering a SNP outside of the recombined left homology arm. Detection of both alleles in the amplicon confirms homozygous AID knock-in. b, Fixed-cell imaging of HaloTag in wild-type cells (left) or in SPEN-degron mESCs (middle) exposed to auxin for 4 hours (right). Cells were labeled with Halo-JF646 before fixation. SPEN-Halo is properly localized to the nucleus, and depleted upon auxin treatment. This experiment was repeated at least twice with similar results. c, Barplot representing the proportion of cells displaying Xist RNA clouds (quantified using RNA FISH) before and after degradation of SPEN (n: number of cells counted, Chi-squared test). d, Violin plot showing X-chromosomal transcript allelic ratios distribution (RNAseq) after 0h dox, 24h dox or 24h dox+auxin treatment in Spen-degron mESCs wild-type cells (horizontal lines denote the median, box limits correspond to upper and lower quartiles, averages of two independent clones shown, n=434 genes, two-sided Student’s t-test). e, RNA FISH for Xist (red) and two X-linked genes: Atrx (grey) and Huwe1 (green), in SPEN-degron mESCs treated with doxycycline only, or in combination with auxin for 24 hours. The proportion of Atrx/Huwe1 monoallelic and biallelic expression among Xist-expressing cells is shown (n: number of cells counted, Chi-squared test). f, Schematic of the control hybrid mouse cross set up for the experiment shown in Fig. 1g-h. g, qPCR quantification of Spen and Xist transcripts in wild-type (n=7) and maternal-zygotic Spen KO (n=5) E3.5 embryos. Bars show mean value. h, Pyrosequencing assay of 3 X-linked transcripts in maternal-zygotic Spen ko (n=5) and WT (n=7) E3.5 embryos (two-sided Student’s t-test). Bars show mean value.