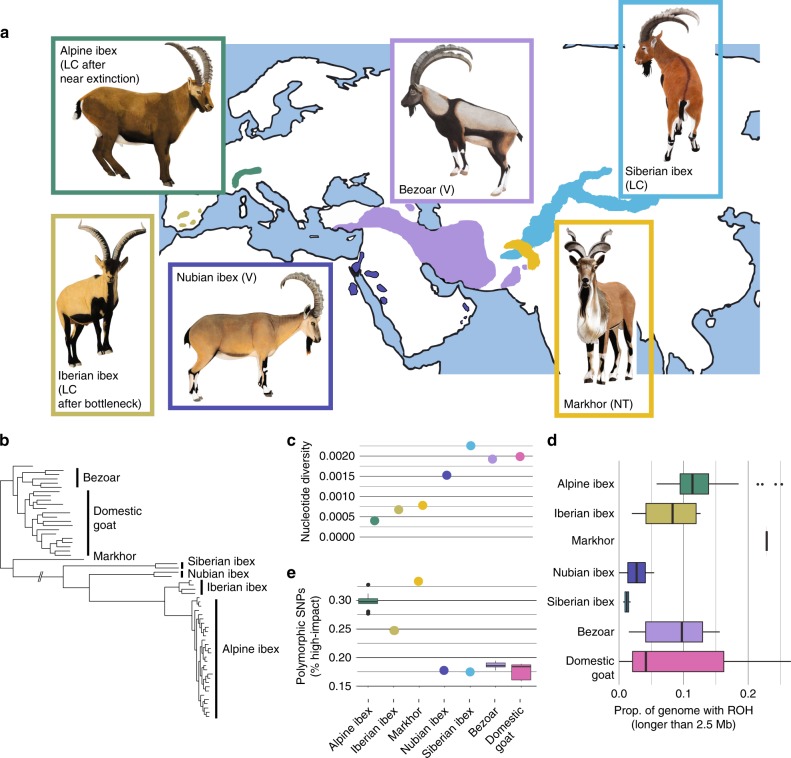
Fig. 1. Genetic diversity and deleterious mutation load of ibex species.
a Geographical distribution and IUCN conservation status of ibex and wild goat species (LC: Least concern, V: Vulnerable, NT: Near threatened39). Sample sizes: Alpine ibex (C. ibex): N = 29, Iberian ibex (C. pyrenaica): N = 4, Bezoar (C. aegagrus): N = 6, Siberian ibex (C. sibirica): N = 2, Markhor (C. falconeri): N = 1, Nubian ibex (C. nubiana): N = 2. b Maximum likelihood phylogenetic analyses, (c) nucleotide diversity, (d) proportion of the genome with runs of homozygosity (ROH) longer than 2.5 Mb and (e) percentage of polymorphic sites within species that segregate highly deleterious mutations. Confidence intervals are based on downsampling to four individuals matching the sample size of Iberian ibex (100 replicates). Boxplots show the median, the 25th and 75th percentiles, Tukey whiskers (median ± 1.5 times interquartile range), and outliers (•). Source data are provided as a Source Data file.