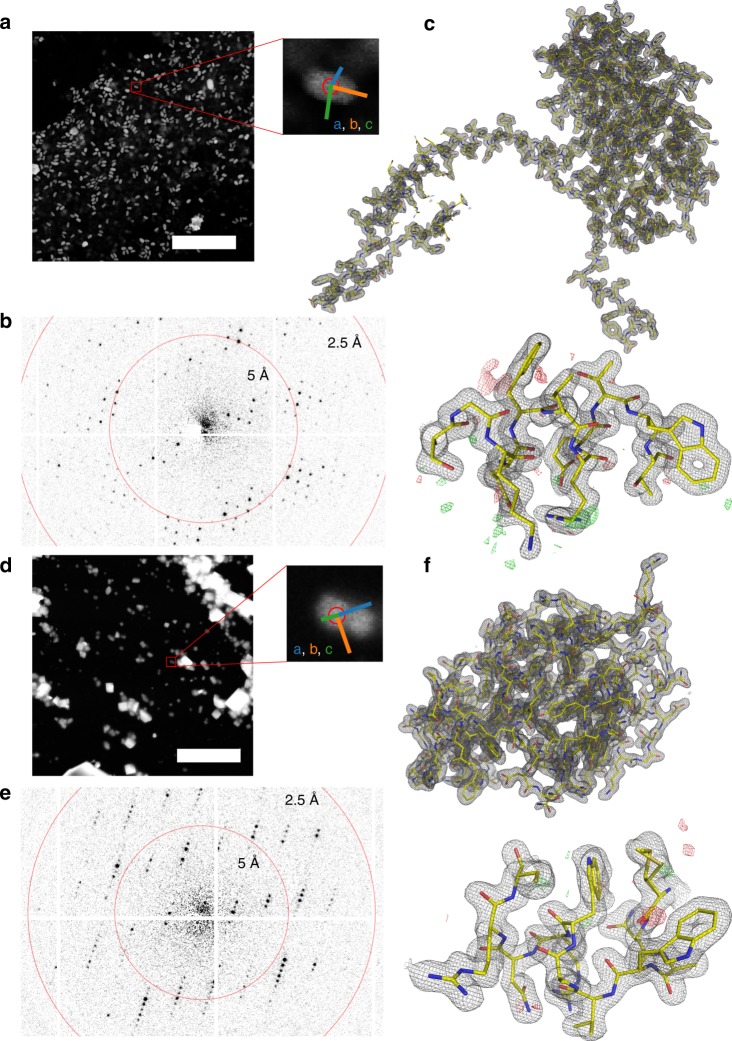
Fig. 2. SerialED results for granulovirus occlusion bodies and lysozyme.
a STEM mapping image of a grid section containing granuloviruses, visible as bright features (scale bar is 5 μm). A zoomed view of a representative virus is shown, where the red circle corresponds to the diffraction nanobeam diameter of ≈110 nm. Colored lines indicate the lattice vectors found after indexing of the diffraction pattern. b Diffraction pattern acquired from the features shown in a. c Obtained structures of granulin; 2Fo−Fc map of the entire structure, and zoom into a randomly chosen region, with Fo−Fc map overlaid. The maps are at 1.55 Å resolution and contoured at ±1σ and ±3σ, respectively. d–f Analogous for lysozyme nanocrystals; maps are at 1.8 Å resolution.