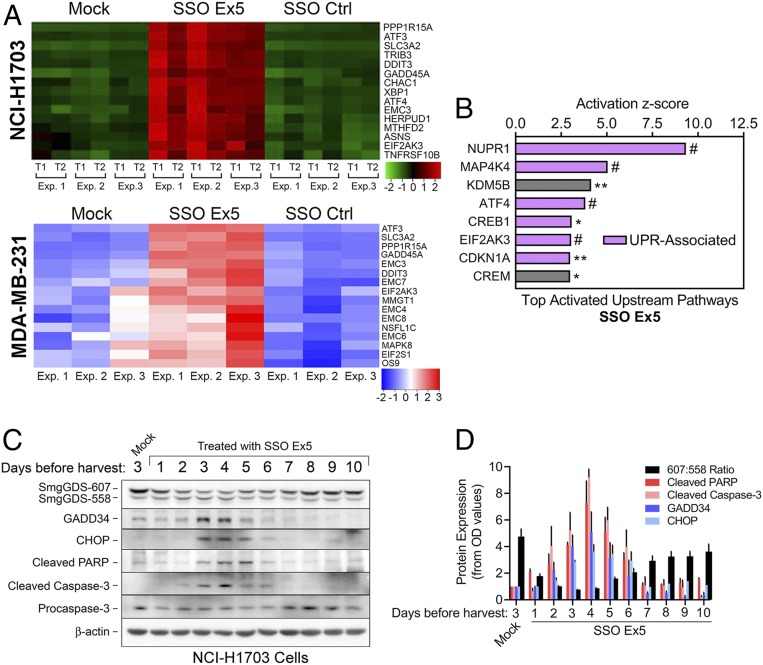
Fig. 5.
SSO Ex5 activates the unfolded protein response. (A) Heatmaps of RNA-seq microarray data display changes in expression of genes involved with ER stress and the UPR that were most significantly altered in NCI-H1703 (Top) and MDA-MB-231 (Bottom) cells treated with mock, SSO Ex5, or SSO Ctrl for 72 h. For the NCI-H1703 heatmap, data were generated in 2 technical replicates (T1 and T2) from 3 independent experiments (Exp. 1, 2, and 3). For the MDA-MB-231 heatmap, data were generated from 3 independent experiments (Exp. 1, 2, and 3). (B) IPA of RNA-seq microarray data from NCI-H1703 cells treated with SSO Ex5 for 72 h identified significant activation of the indicated upstream signaling pathways. Purple bars indicate pathways that are directly associated with the canonical UPR, or pathways that have been reported to be indirectly associated with ER stress and the UPR. Pathways are ordered by their corresponding z score, and labeled for IPA gene overlap significance (*P < 0.05, **P < 0.01, #P < 0.0001). (C) NCI-H1703 cells were treated once with mock or SSO Ex5, and lysates were collected at the indicated days after treatment for immunoblot analysis of apoptotic and UPR factors. (D) Densitometric analysis of immunoblots from C was performed by normalizing the OD values of cleaved PARP, cleaved caspase-3, GADD34, and CHOP to β-actin. Values were then normalized to mock-treated cells and plotted, along with the 607:558 ratio. Results are presented as mean ± SEM (n = 3).