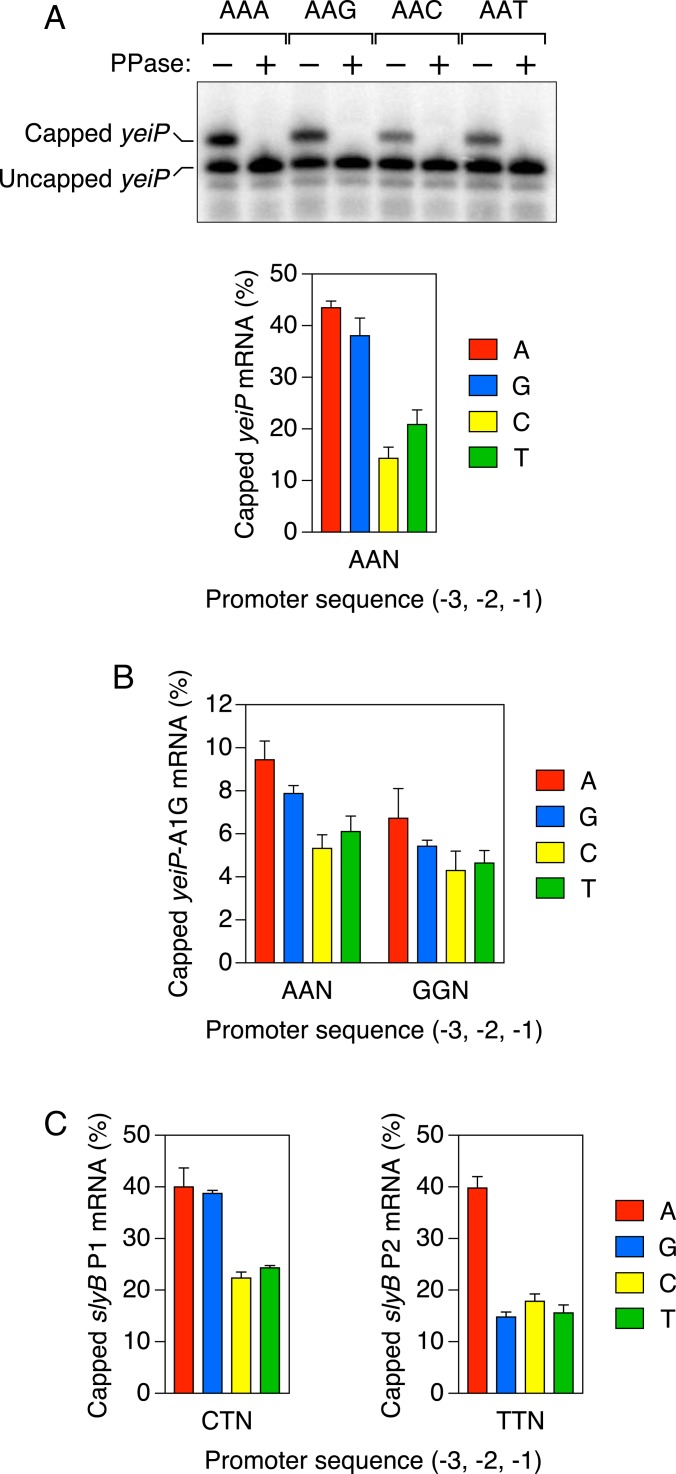
Fig. 2.
Promoter-dependent capping of mRNA in E. coli. (A) Percentage of yeiP transcripts that are Np4-capped in unstressed ∆apaH cells. (Top) Northern blot. Total RNA was extracted from unstressed ∆apaH cells that lacked the chromosomal yeiP gene and instead contained a plasmid-borne yeiP gene in which the sequence of the coding strand at positions −3, −2, and −1 of the promoter had been changed to AAN, where N = A, G, C, or T. The RNA was cleaved site-specifically 69 nt from the yeiP 5′ end with a 10-23 deoxyribozyme (24) so as to generate a 5′-terminal fragment bearing a 2′,3′ cyclic phosphate rather than a vicinal diol at its 3′ end, and samples containing similar amounts of uncapped yeiP RNA were then subjected to electrophoresis on a boronate gel, blotted, and probed with radiolabeled yeiP DNA (26). Equivalent RNA samples that had been decapped in vitro with RppH (PPase) were analyzed in parallel. (Bottom) Graph. For each promoter, the percentage of yeiP transcripts that were capped was calculated from the mean of four or five biological replicates. Error bars represent 1 SD. P ≤ 0.001 for each AAPu vs. AAPy comparison, where Pu = A or G and Py = C or T. (B) Percentage of yeiP-A1G transcripts that are Np4-capped in unstressed ∆apaH cells. Each value corresponds to the mean of five biological replicates for yeiP-A1G mRNA synthesized by transcription from promoter variants in which the sequence of the coding strand at positions −3, −2, and −1 had been changed to AAN or GGN, where N = A, G, C, or T. Error bars represent 1 SD. P ≤ 0.001 for each AAPu vs. AAPy comparison, where Pu = A or G and Py = C or T; P ≤ 0.025 for each GGPu vs. GGPy comparison; P ≤ 0.05 for each cognate AAN vs. GGN comparison. (C) Percentage of slyB P1 and slyB P2 transcripts that are Np4-capped in unstressed ∆apaH cells. Each value represents the mean of five biological replicates for slyB P1 mRNA (Left) or slyB P2 mRNA (Right) synthesized by transcription from promoter variants in which the sequence of the coding strand at position −1 had been changed to A, G, C, or T. Error bars represent 1 SD. The value for slyB P1 bearing an A at position −1 represents a lower limit on the actual percentage of capped transcripts, as this mutation adventitiously created a third slyB promoter whose capped transcript comigrated with the uncapped form of slyB P1 mRNA. P ≤ 0.001 for each CTPu vs. CTPy comparison and each TTA vs. TTPy comparison, where Pu = A or G and Py = C or T.