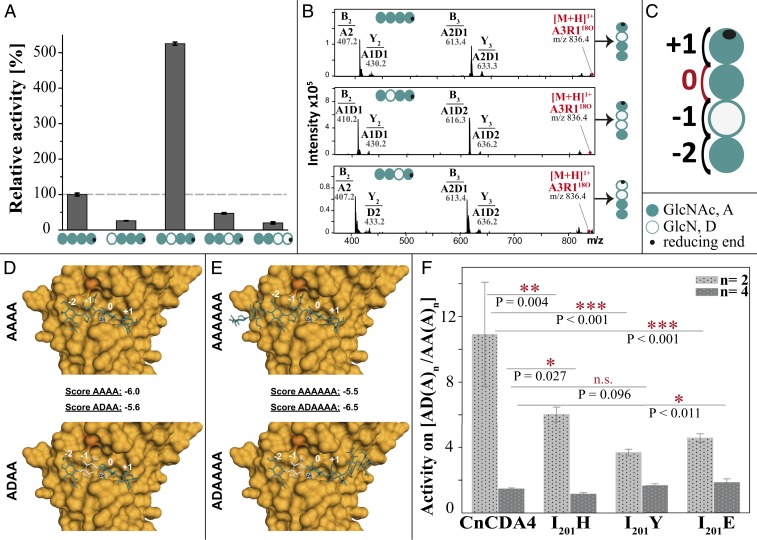
Fig. 2.
Regioselectivity and regiospecificity of CnCda4 against partially acetylated chitosan tetramers and hexamers. (A) Relative activity of CnCda4 against five tetrameric substrates differing in DA and PA, as determined by quantitative MS1 (performed with four independent enzyme batches of CnCda4; values are means ± SD). (B) MS2 spectra of the deacetylation products of A4, ADAA, and AADA after fragmentation, showing the fragment masses and composition of b-ions (nonreducing end) and y-ions (reducing end). (C) Model representing the binding preferences of CnCda4 at four subsites (−2, −1, 0, +1) where subsite 0 is the catalysis site. (D) Homology model of CnCda4 (based on PDB ID code 2IW0), with Zn2+ (gray) and I201 in dark orange, showing docking results for AAAA and ADAA in the active site. Subsites −2, −1, 0, and +1 are indicated in white. Docking scores are given for all substrates. (E) The same homology model showing docking results for AAAAAA and ADAAAA. (F) Activity ratio of wild-type CnCda4 and three mutants against partially deacetylated AD(A)n vs. fully acetylated AA(A)n, where n = 2 for tetrameric substrates and n = 4 for hexameric substrates (values are means of four replicates each ±SD).