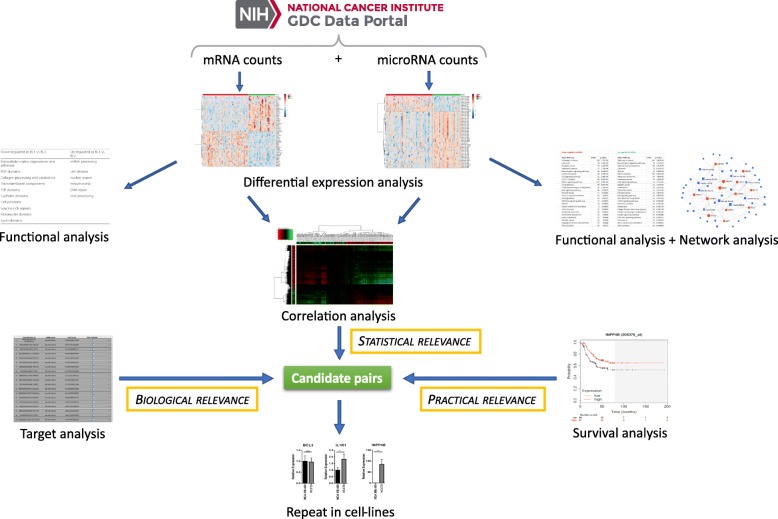
Fig. 9.
Integrative analysis approach. Raw RNA counts from GDC database were processed in differential expression analysis. Differentially expressed RNAs were further inspected via functional analysis and network analysis (for microRNAs) to confirm that the significant differences are cancer-related. Afterwards, correlation analysis, target analysis and survival analysis were jointly applied to the differentially expressed RNAs to select the best candidates that could affect the difference between BL1 and BL2 subtypes and their outcomes. The candidates were then verified in BL1 and BL2 cell lines