Figure 1.
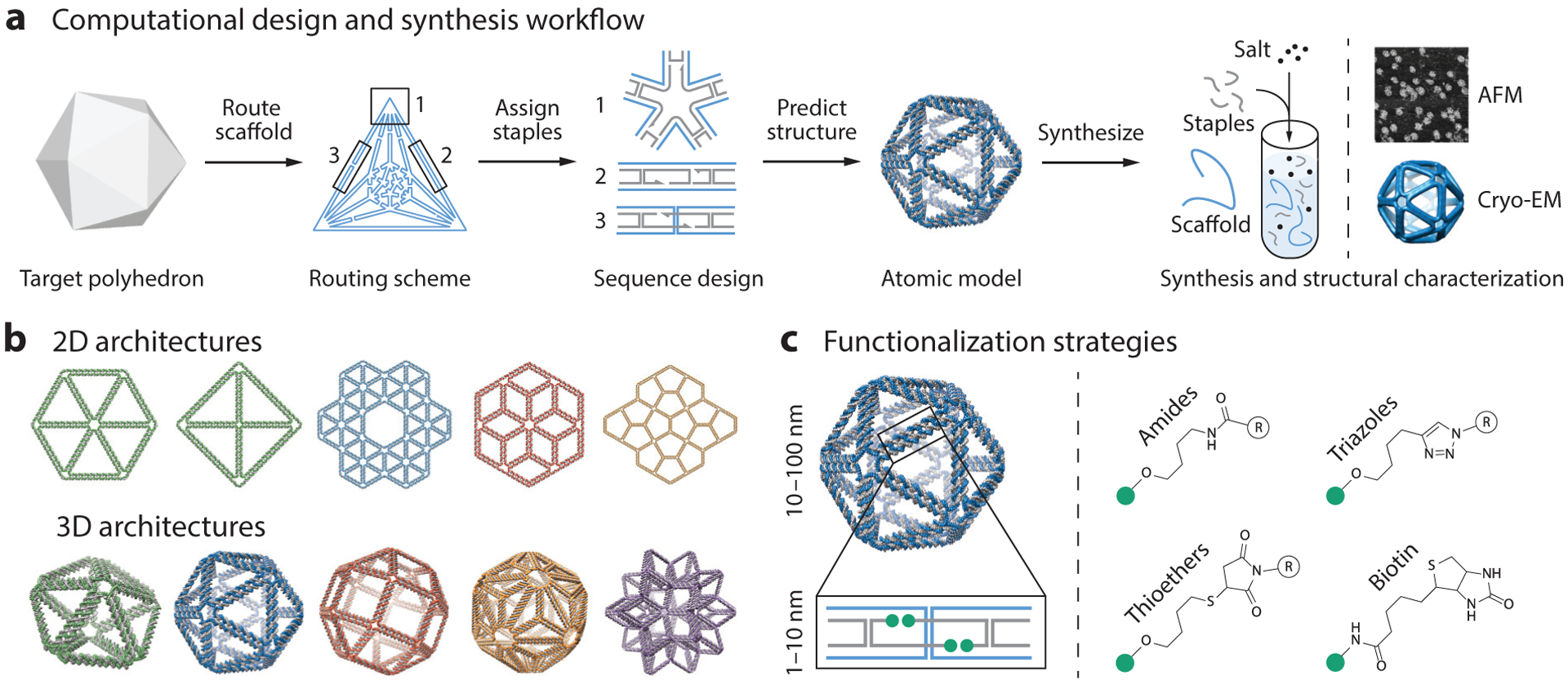
Design and synthesis of wireframe scaffolded DNA origami nanostructures for biophysical research. (a) This panel shows the workflow for designing DNA origami objects using the software DAEDALUS (186). First, a target polyhedron is used as input to the algorithm. DAEDALUS then automatically routes the scaffold through every edge and assigns the staples needed to fold the target object. An atomic model is also generated to allow researchers to visualize the object and design molecular functionalization patterns. Using the computationally designed set of oligonucleotide sequences, the DNA nanoparticle is self-assembled via thermal annealing with an excess of staples over scaffold, which is typically based on the M13 phage or a synthetic sequence. Folding is characterized using agarose gel electrophoresis and structural analysis using atomic force microscopy (AFM), transmission electron microscopy (TEM), or 3D cryo-electron microscopy (cryo-EM). Adapted with permission from Reference 186. (b) A vast number of different wireframe architectures of arbitrary shape can now be designed from the top down using algorithms such as DAEDALUS (186; http://daedalus-dna-origami.org/) and PERDIX (86; http://perdix-dna-origami.org/). Adapted with permission from References 86 and 186. (c) Biomolecular functionalization on wireframe objects can be controlled with single-molecule precision. Strategies for the functionalization of staples at 3′ or 5′ locations include hybridization of single-stranded DNA overhangs with complementary locked nucleic acid or peptide nucleic acid sequences and direct, covalent chemical modifications.
