Figure 2.
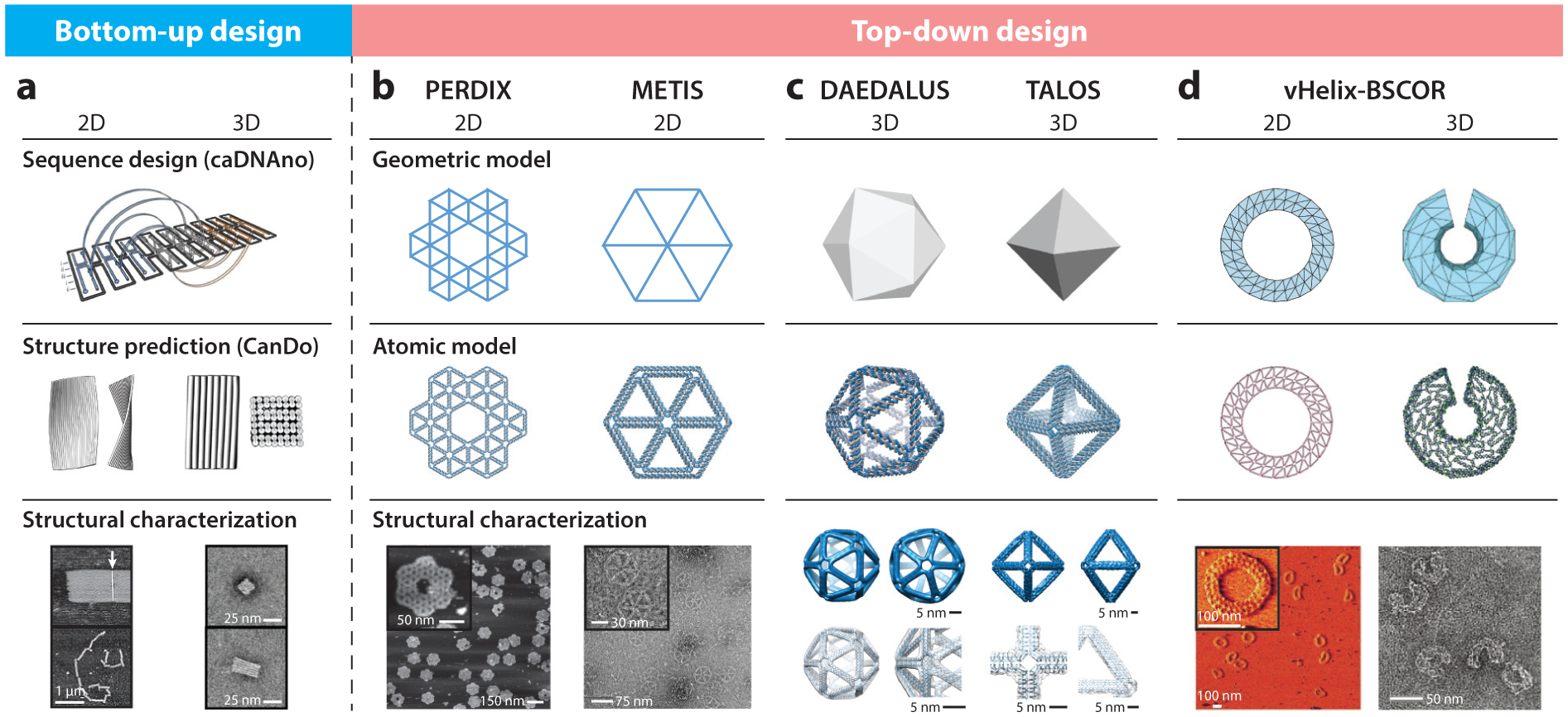
Computational design strategies for scaffolded DNA origami nanostructures. Following the early development of the bottom-up sequence design program caDNAno for rectilinear, brick-like scaffolded DNA origami assemblies, several approaches for the top-down, fully automated design of wireframe scaffolded DNA origami objects were developed, including PERDIX, METIS, DAEDALUS, TALOS, and vHelix-BSCOR. (a) Sequence designs from caDNAno can be imported into the online software CanDo to predict three-dimensional (3D) solution structures. Adapted with permission from References 42, 43, 92, and 143. (b) Both PERDIX and METIS enable the design of any free-form 2D geometry using either DX- or honeycomb-based edges, respectively. Adapted with permission from Reference 86. (c) DAEDALUS and TALOS program arbitrary 3D polyhedral geometries using DX- or honeycomb-based edges, respectively. Adapted with permission from References 186 and 85. (d) vHelix-BSCOR enables the semiautomated top-down design of both 2D and 3D wireframe objects using predominantly single-helix edge architectures (right). Adapted with permission from References 17 and 18. Structural characterization of the target objects is typically achieved using atomic force microscopy for 2D assemblies (a, bottom left, b, bottom left, and d, bottom left, where the white arrow indicates blunt-end stacking of Rothemund rectangles in panel a), cryo-electron microscopy with 3D reconstruction (c, bottom left and right) or transmission electron microscopy (a, bottom right; b, bottom right; d, bottom right)
