Mycoplasma bovis is a destructive pathogen that causes large economic losses in rearing cattle for beef and dairy worldwide. M. bovis causes suppression of and evades the host immune response; however, the mechanisms of host immune function involved in M. bovis mastitis have not been elucidated. The purpose of this study was to elucidate the characteristics of the bovine immune response to mycoplasmal mastitis. We evaluated the responsiveness of the bovine mammary gland following infusion of M. bovis.
KEYWORDS: mycoplasma, cattle, veterinary immunology
ABSTRACT
Mycoplasma bovis is a destructive pathogen that causes large economic losses in rearing cattle for beef and dairy worldwide. M. bovis causes suppression of and evades the host immune response; however, the mechanisms of host immune function involved in M. bovis mastitis have not been elucidated. The purpose of this study was to elucidate the characteristics of the bovine immune response to mycoplasmal mastitis. We evaluated the responsiveness of the bovine mammary gland following infusion of M. bovis. Somatic cell counts and bacterial counts in milk from the infected quarter were increased. However, the proliferation of peripheral blood mononuclear cells (blood MNCs) and mononuclear cells isolated from M. bovis-stimulated mammary lymph nodes (lymph node MNCs) did not differ from that in the unstimulated cells. Transcriptome analysis revealed that the mRNA levels of innate immune system-related genes in blood MNCs, complement factor D (CFD), ficolin 1 (FCN1), and tumor necrosis factor superfamily member 13 (TNFSF13) decreased following intramammary infusion of M. bovis. The mRNA levels of immune exhaustion-related genes, programmed cell death 1 (PD-1), programmed cell death-ligand 1 (PD-L1), lymphocyte activation gene 3 (LAG3), and cytotoxic T-lymphocyte-associated protein 4 (CTLA4) of milk mononuclear cells (milk MNCs) in the infected quarter were increased compared with those before infusion. Increase in immune exhaustion-related gene expression and decrease in innate immune response-related genes of MNCs in quarters from cows were newly characterized by M. bovis-induced mastitis. These results suggested that M. bovis-induced mastitis affected the immune function of bovine MNCs, which is associated with prolonged duration of infection with M. bovis.
INTRODUCTION
Mycoplasmas, bacteria of the Mollicutes class, do not have a cell wall, are widespread in nature, and infect eukaryotes (1). Mycoplasma bovis is a destructive pathogen of beef and dairy cattle worldwide (2, 3) that is known to be a major contributing factor to the occurrence of mastitis, pneumonia, and arthritis (2, 4), which all contribute to large economic losses on dairy farms (2, 5). M. bovis infection leads to calf mortality, weight loss in surviving calves, and a decline in milk production in dairy cows (2, 6). Mycoplasma mastitis, particularly intramammary infection with M. bovis, increases somatic cell counts (SCCs; 107 to 109 cells/ml) in milk, as do other pathogens causing mastitis (7–9). M. bovis-induced mastitis causes swelling and induration of the udder following a marked decrease of milk yield (3). M. bovis-induced mastitis is also known to have a long infection period and shows a subclinical type of mammary infection (10). It has been reported that M. bovis-induced mastitis causes a sustained increase in SCCs and proinflammatory cytokines in milk (9). However, the sustained inflammatory response is not strong enough to eradicate M. bovis from the mammary gland (9). The major cell population present in mastitis comprises neutrophils (>90%), and the influx of neutrophils into the mammary gland is mediated by mononuclear cells (MNCs) (11). Although subpopulations of MNCs are of various types and have various functions (12), the trafficking of different lymphocyte subpopulations in M. bovis-induced mastitis is not fully understood.
M. bovis is thought to evade the host immune response, and our previous study showed that M. bovis evades bovine neutrophil extracellular traps (13). M. bovis exhibits immunosuppressive characteristics that inhibit the proliferation of lymphocytes in response to mitogens (14–16). In a long-period infection, immunosuppressive factors, including programmed cell death 1 (PD-1), lymphocyte activation gene 3 (LAG3), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), T cell immunoglobulin, and mucin-domain containing-3 (Tim3), are expressed on lymphocytes (17). These proteins bind to their respective ligands to induce immune exhaustion of the effector cells (17). These immune exhaustion-related proteins induce a marked reduction in cell proliferation and cytotoxic activity (18, 19). However, the underlying mechanisms of the immunosuppressive host immune response of M. bovis-induced mastitis have not been elucidated.
The purpose of this study was to elucidate the characteristics of the host immune response to mycoplasmal mastitis. Furthermore, we aimed to evaluate the response of the bovine mammary gland after infusion of M. bovis.
RESULTS
Quantification of the SCC and bacterial counts in milk.
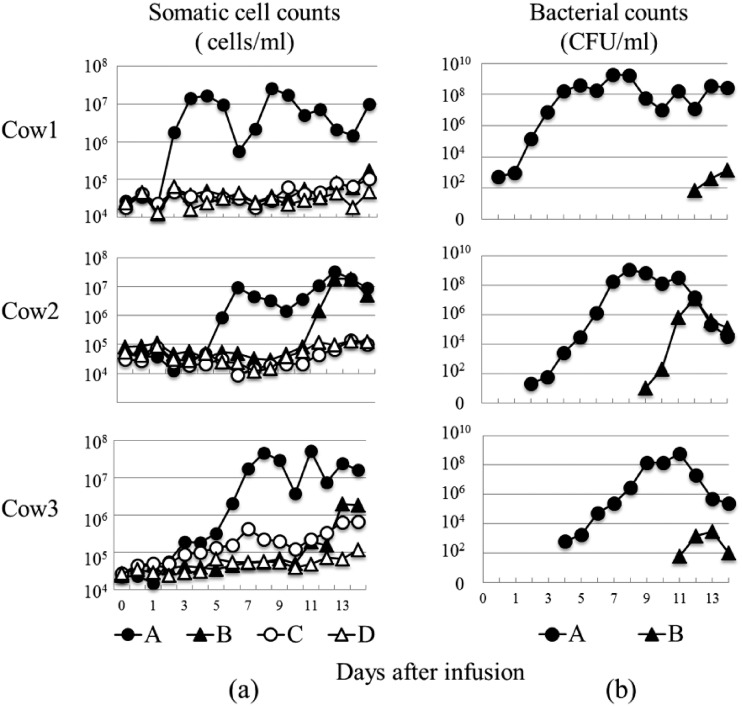
The SCCs of each cow following intramammary infusion of M. bovis are shown in Fig. 1a. The SCCs of the infected quarter at day 2 to 5 increased sharply in comparison with the SCCs of day 1 in three cows. The SCCs of the infected quarter of each cow at days 8 to 11 peaked at >107 cells/ml. The SCCs of B quarter at days 9 to 11 increased compared with that at day 1, and the SCCs of B quarter at day 14 (cow1), day 12 (cow2), and day 13 (cow3) peaked at >105 (cow1), >107 (cow2), and >106 (cow3) cells/ml, respectively. The SCCs of C and D quarters tended to increase until day 14 postinfusion of M. bovis, but the SCC was <3.0 × 105 cells/ml (except for quarter C in cow3 at day 7 and days 12 to 14). The bacterial counts following intramammary infusion of M. bovis are shown in Fig. 1b. The bacterial counts of the infusion quarter at day 7 to 11 peaked at 108 to 109 CFUs/ml in three cows. M. bovis was detected in all B quarters in three cows, and the bacterial counts of M. bovis were >103 (cow1 and cow3) and >107 (cow2) CFU/ml.
FIG 1.
SCC and bacterial counts following intramammary infusion of M. bovis. (a) SCC (×104 cells/ml) and (b) bacterial counts (CFU/ml) evaluated at days 0, 0.5, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, and 14 postinoculation in the M. bovis inoculation quarter (A, left forequarter), PBS inoculation quarter (D, right rear quarter), and other quarters (B, left rear quarter; C, right forequarter) in three cows.
Surface marker analysis of blood MNCs and milk MNCs.
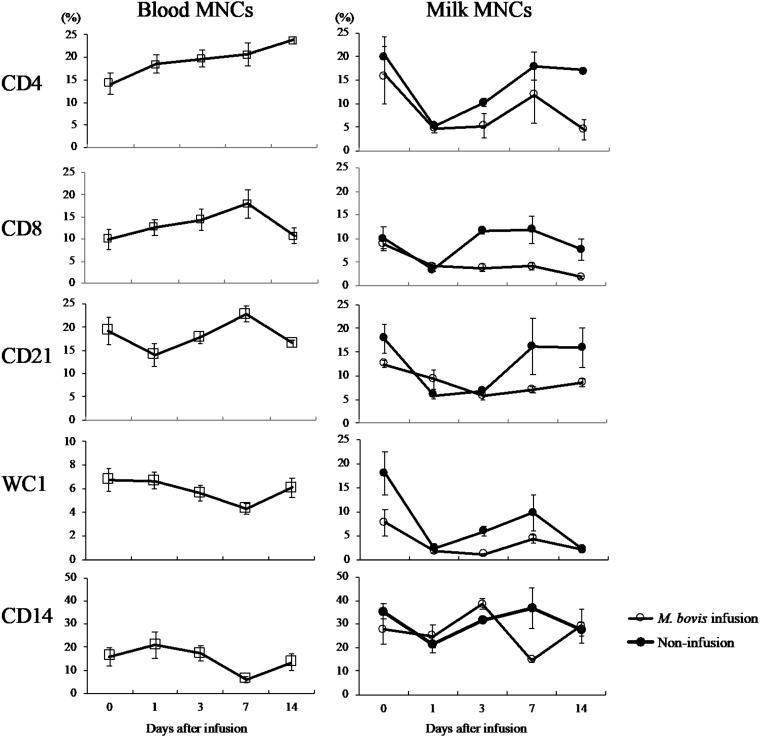
The surface marker analysis of blood MNCs and milk MNCs is shown in Fig. 2. The average ratios of CD4-, CD8-, CD21-, WC1-, and CD14-positive cells in the blood MNCs were not significantly altered. However, the average ratio of CD4-positive cells in the blood MNCs was increased until day 14 compared with that at day 0. The average ratios of CD4-, CD8-, and CD21-positive cells in the milk of the infected quarter at days 7 to 14 were higher than those of the noninfusion quarter (D quarter). The average ratios of WC1 (days 1, 3, and 7)- and CD14 (day 7)-positive cells in the milk of the infected quarter were higher than those of the noninfusion quarter, D quarter.
FIG 2.
Surface marker analysis of blood MNCs and milk MNCs following intramammary infusion of M. bovis. Effect at days 0, 1, 3, 7, and 14 following intramammary infusion of M. bovis on the populations of blood MNCs and milk MNCs in the M. bovis inoculation quarter (left forequarter) and noninfusion quarter (right rear quarter) as determined by flow cytometry. Data are presented as the mean ± SEM of the results from three cows. Percentages of cells in blood MNCs and milk MNCs gate positive for CD4, CD8 CD21, WC1, and CD14.
Proliferation of MNCs stimulated with M. bovis.
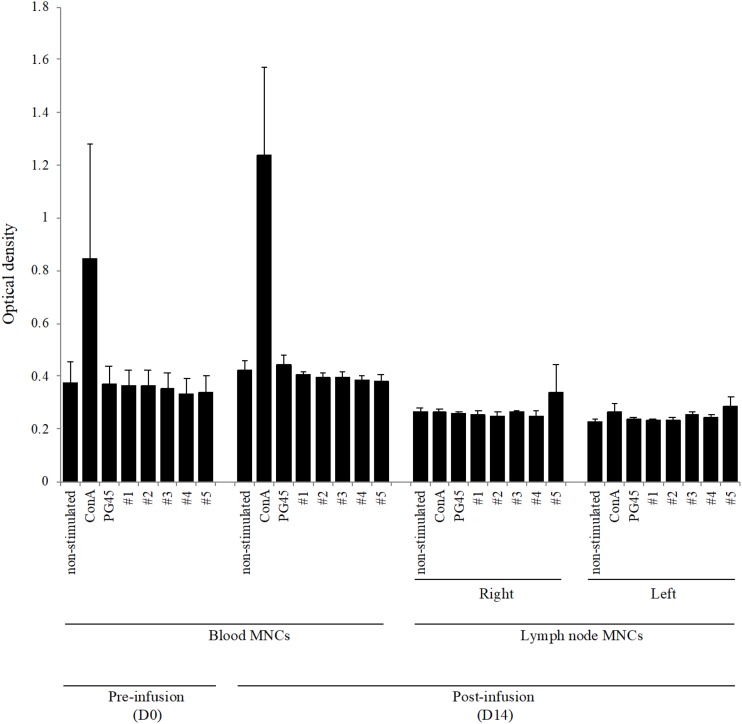
The proliferation of blood MNCs and lymph node MNCs stimulated with concanavalin A (ConA) and M. bovis at day 0 (only blood MNCs) and day 14 is shown in Fig. 3. The proliferation of blood MNCs stimulated with ConA at day 14 was higher than that at day 0; however, M. bovis did not induce changes in cell proliferation (both PG45 and five wild-type strains). The lymph node MNCs stimulated with M. bovis did not exhibit a change in proliferation compared with that of the nonstimulated cells.
FIG 3.
Proliferation of blood MNCs and lymph node MNCs stimulated with M. bovis or ConA. Bovine blood MNCs and lymph node MNCs were incubated with live M. bovis (multiplicity of infection of 100; PG45 strain and five wild-type strains) or ConA. The optical density (OD) values are expressed as the mean ± SE from the results in three cows.
Microarray analysis.
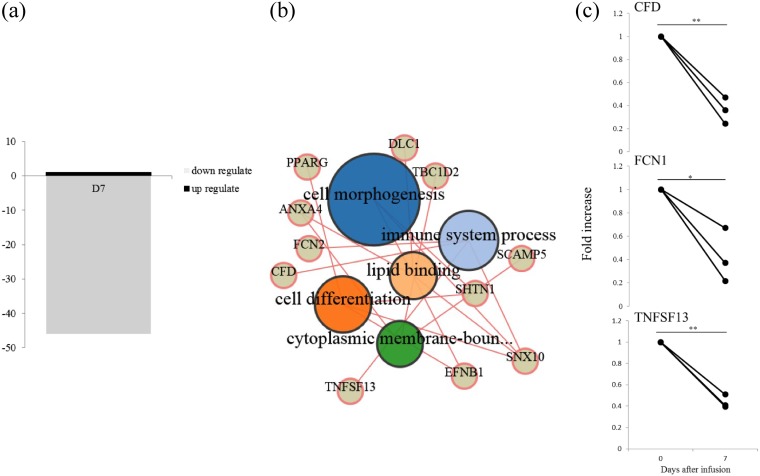
Statistical analysis revealed that at day 7, following intramammary infusion of M. bovis, the expression levels of 47 genes in the blood MNCs were significantly altered (P < 0.025 with a fold increase of >2) compared with those at day 0 (Fig. 4a; Table S1 in the supplemental material). Only the gene expression of transglutaminase 3 (TGM3) in blood MNCs was significantly increased, whereas the expression levels of the remaining 46 genes were significantly decreased. Gene ontology (GO) terms were identified as cell morphogenesis and immune system process (Fig. 4b; Table S2 in the supplemental material). For validation of this result, genes associated with the immune system were quantified using real-time PCR (Fig. 4c). The mRNA expression levels of complement factor D (CFD), ficolin 1 (FCN1), and tumor necrosis factor superfamily member 13 (TNFSF13) of the blood MNCs at day 7 were significantly (P < 0.01) decreased compared with those at day 0.
FIG 4.
Microarray analysis in blood MNCs following intramammary infusion of M. bovis. Blood MNCs evaluated at days 0 and 7 postintramammary infusion of M. bovis in three cows. (a) Number of significantly downregulated or upregulated mRNAs; (b) gene ontology term enrichment analysis of genes recognized as significantly different in blood MNCs following intramammary infusion of M. bovis; (c) quantification of mRNA expression by microarray analysis; significant difference at *P < 0.05 and **P < 0.01 compared with day 0.
Immune exhaustion-related gene expression.
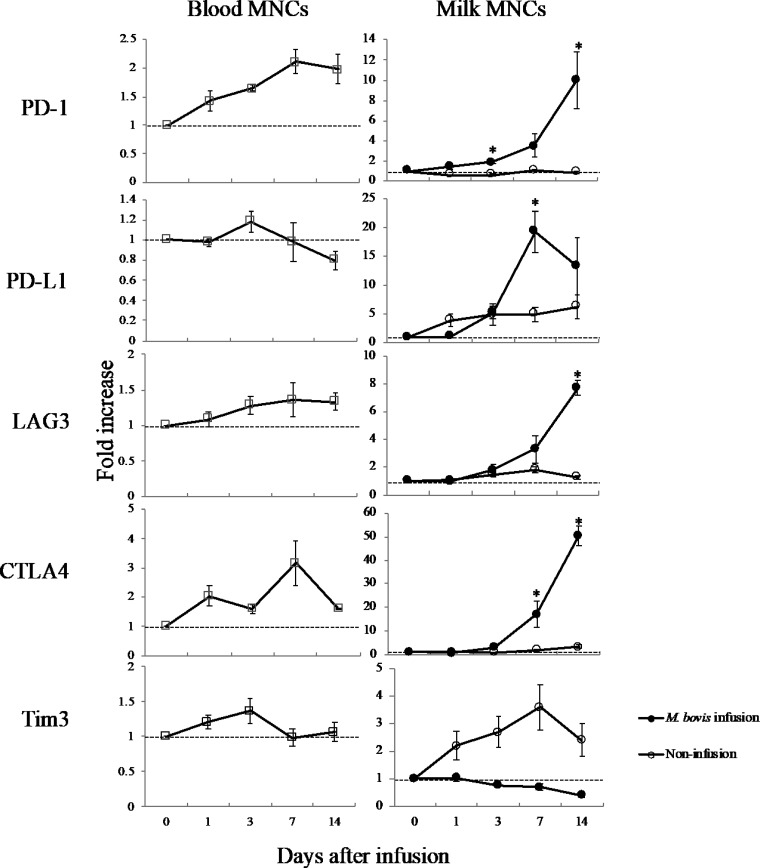
The mRNA expression levels of PD-1, PD-1 ligand 1 (PD-L1), lymphocyte activation gene 3 (LAG3), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), and T cell immunoglobulin and mucin-domain containing-3 (Tim3) in blood MNCs and milk MNCs following intramammary infusion of M. bovis were evaluated (Fig. 5). The mRNA expression levels of PD-1, LAG3, and CTLA4 in the blood MNCs following intramammary infusion of M. bovis were higher than those at day 0. The mRNA expression levels of PD-1, PD-L1, LAG3, and CTLA4 of milk MNCs in the M. bovis-infected quarter were increased compared with those at day 0; however, the mRNA expression levels of PD-1, LAG3, and CTLA4 in the milk MNCs of the noninfusion quarter (D quarter) were unchanged. The mRNA expression levels of PD-1 (at days 3 and 14), PD-L1 (at day 7), LAG3 (at day 14), and CTLA4 (at days 7 and 14) of milk MNCs were significantly (P < 0.05) increased compared with those at day 0.
FIG 5.
mRNA expression of immune exhaustion-related genes following intramammary infusion of M. bovis. Blood MNCs and milk MNCs from the M. bovis inoculation quarter (A, left forequarter) and noninfusion quarter (D, right rear quarter), following intramammary infusion of M. bovis, were evaluated on days 0, 1, 3, 7, and 14. The mRNA expression levels of PD-1, PD-L1, LAG3, CTLA4, and Tim3 were determined by real-time PCR and expressed as a fold increase. The data are expressed as the mean ± SE from the results in three cows; significant difference at *P < 0.05 compared with day 0.
DISCUSSION
The present study characterized the response of the immune system to M. bovis-induced mastitis, particularly the effects on immune function in MNCs during M. bovis-induced mastitis. Following intramammary infusion of M. bovis, the maximal bacterial counts of M. bovis in milk were 108 to 109 CFU/ml. Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa induce counts of 103 to 105 CFU/ml in milk following their intramammary infusion (7, 8). The results of this study showed that bacterial counts in milk from the M. bovis-infected quarter were 103 to 106 times higher than those from other pathogens. Although the specific mechanisms for this increased infection are unknown, one possible explanation is the evasion of M. bovis from the immune response of the mammary gland. In this study, M. bovis was detected not only in the A quarter (infused quarter) but also in the B quarter (noninfused quarter). These results were consistent with a previous report that M. bovis can spread to nonchallenged quarters (9). The maximal SCC reached a peak of 37.0 × 106 ± 3.8 × 106 cells/ml in the infused quarter. Bannerman et al. (7, 8) reported that the SCC level of milk obtained from bacterium-infused quarters of E. coli was 44.9 × 106 ± 5.0 × 106 cells/ml; that from S. aureus, 32.1 × 106 ± 5.9 × 106 cells/ml; and that from P. aeruginosa, 62.9 × 106 ± 6.4 × 106 cells/ml. From these results, the highest level of SCCs following the intramammary infusion of M. bovis was similar to that of E. coli and S. aureus infusions.
In the present study, the major cells of recruitment in the milk of M. bovis-induced mastitis were neutrophils, as with other causes of mastitis (11). Although MNCs serve an important role in the influx of neutrophils to the mammary gland or are associated with excessive inflammatory response (11), characterization of the surface marker of MNCs has not been fully clarified in M. bovis-induced mastitis. In the present study, it was first shown that CD4- and CD8-positive cells at day 3, 7, and 14 postinfusion of M. bovis were marginally increased compared with those in the D quarter (noninfusion quarter). CD4-positive T cells enhance humoral and cell-mediated immunity. In addition, CD8-positive T cells are associated with cell-mediated immunity and the T cell-mediated recruitment of neutrophils during bacterial infection (20). We showed that the numbers of CD21-positive cells in milk at days 3, 7, and 14 postinfusion of M. bovis tended to be higher than those in the D quarter. CD21-positive cells are capable of antigen presentation (21), secretion of cytokines (22), and differentiation into plasma cells that produce immunoglobulins (23). It has been reported that CD4-, CD8-, and CD21-positive cells in milk are significantly increased in S. aureus- or E. coli-induced mastitis (24–26). Our results suggest that the CD4-, CD8-, and CD21-positive cells in the milk MNCs are weakly affected by M. bovis-induced mastitis compared with E. coli- or S. aureus-induced mastitis.
Of note, the stimulation of M. bovis in MNCs isolated from mammary lymph nodes and blood MNCs following intramammary infusion of M. bovis did not induce cell proliferation. It has been reported that blood MNC proliferation is inhibited by M. bovis infection (14–16) and that M. bovis induces the colonization of other pathogenic bacteria (2, 27). Our results suggest that effects on immune function in M. bovis infection may prevent proliferation.
The expression levels of genes associated with the innate immune response were significantly decreased in the transcriptome analysis of blood MNCs at day 7. GO enrichment analysis suggests that M. bovis-induced mastitis influences the suppression of cell morphogenesis, which is related to the cell migration and immune system process. Two genes associated with complement activation were significantly decreased; FCN1 is associated with the lectin pathway and CFD is associated with another pathway (28, 29). TNFSF13, associated with the proliferation of B cells, was also significantly decreased (30). Although further studies are required, our results indicate that M. bovis-induced mastitis influenced inhibition of the host innate immune response. In addition, the mRNA expression levels of PD-1, LAG3, and CTLA4 of blood MNCs were higher following the intramammary infusion of M. bovis. The mRNA expression levels of PD-1, PD-L1, LAG3, and CTLA4 in milk MNCs were also increased. These immune checkpoint-related genes are known to be associated with T cell exhaustion (17), inducing marked reductions in cell proliferation and cytotoxic activity (18, 19). Our results suggest that M. bovis induced immune exhaustion following the downregulation of cell proliferation. This is the first report to demonstrate that M. bovis can trigger immune exhaustion in M. bovis-induced mastitis. M. bovis affects the cell function and gene expression of bovine MNCs in M. bovis-induced mastitis, which may contribute to a long period of infection and lead to severe mastitis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study were M. bovis (PG45, ATCC 25523 and five wild-type strains). M. bovis was grown in modified PPLO medium (Kanto Kagaku, Tokyo, Japan) at 37°C for 48 h. M. bovis bacteria were obtained by centrifugation (16,000 × g for 40 min) and were then washed with phosphate-buffered saline (PBS). The bacteria were then suspended in PBS to a cell density of 108 CFU/ml, and the suspension was stored at −70°C until used.
Animals.
Three Holstein cows with negative bacteriological culture in milk and SCCs of <200,000 cells/ml at quarter level were used in this study. The SCC was evaluated by the Fossomatic 90 instrument (Foss Electric, Denmark).
Intramammary challenge of M. bovis.
The experimental protocol was approved by the Institutional Animal Care and Use Committee of Rakuno Gakuen University. The left forequarter (A quarter) was infused with ∼50,000 CFUs (10,000 CFUs/ml in 5 ml of sterile PBS) of M. bovis. PBS was infused in the right rear D quarter (as noninfusion quarter). A remnant of the 5-ml intramammary challenge solution was added to modified PPLO agar plates (Kanto Kagaku) for counting the CFU/ml of the M. bovis culture.
Milk and blood sampling.
Milk samples from all four quarters (A, left fore; B, left rear; C, right fore; D, right rear) were tested for SCC on day 0 preinfection control and day 0.5 and days 1 to 14 postinfection from three cows. These milk samples were collected using aseptic techniques and cultured for bacteria, including Mycoplasma spp., at day 0 preinfection control and day 0.5 and days 1 to 14 postinfection. M. bovis in milk was determined as CFU/ml by plating 100 μl of milk onto PPLO modified agar plates (Kanto Kagaku). Milk from highly infected quarters was serially diluted (10-fold) in PBS prior to plating. Following incubation of 1 week at 37°C and 5% CO2, CFU/ml was calculated. PCR for the detection of M. bovis in cultured broth was performed according to the method described by Higuchi et al. (31). For assessing the condition of the cows, rectal temperature, heart rate, and respiratory rate were measured (see Fig. S1 in the supplemental material), and blood was collected for hematological examination, biochemical examination, and peripheral white blood cell counts at day 0 preinfection control and days 1, 3, 7, and 14 postinfection from three cows (see Table S3a to c in the supplemental material).
Blood MNCs and milk MNCs.
Blood samples of 70 ml each were collected from cows using evacuation tubes containing sodium heparin (Terumo, Tokyo, Japan). Milk samples of 1,000 ml (∼107 cells as SCC) were collected into tubes from the A and D quarters. The cells were isolated by centrifugation on a Ficoll-Conray gradient, with specific gravity of 1.080, as previously described (32). The cells were separated by centrifugation (300 × g for 30 min) and were transferred to a sterilized tube (Becton, Dickinson, Tokyo, Japan). The viability of blood MNCs was assessed using an AO/PI cell viability kit (Logos Biosystems, Gyunggi, South Korea) and Luna-FLTM (Logos Biosystems) according to the manufacturer’s protocols.
Surface marker analysis of blood MNCs and milk MNCs.
The cells were adjusted to 106 cells/ml in PBS and surface stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (Bio-Rad clone CC8; for CD4-positive T cell marker), CD8 (Bio-Rad clone CC63; for CD8-positive T cell marker), CD14 (Bio-Rad clone M-M9; for monocyte/macrophage marker), CD21 (Bio-Rad clone CC21; for B cell marker), and WC1 (Bio-Rad clone CC15; for γδ T cell marker) antibody. Following incubation for 15 min in a dark room, the cells were centrifuged (500 × g for 5 min at 4°C), then washed in PBS, and fixed and permeabilized with 1% paraformaldehyde (Polysciences, USA). Data were acquired using the BD FACSVerse (BD Biosciences, USA) system and analyzed on a FACScalibur flow cytometer using CellQuest software (BD Biosciences). Ten thousand events per sample were collected for blood MNCs and 5,000 events per sample were collected for milk MNCs. Data were analyzed for gated MNCs based on the forward- and side-light scatter characteristics, as previously reported (33).
Proliferative response.
Following sacrifice, the bovine mammary lymph nodes (left and right) were immediately collected in cold RPMI 1640 medium (Sigma-Aldrich, Tokyo, Japan), cut using a surgical knife, pulverized, and washed with RPMI 1640 medium. The viability of MNCs was assessed using an AO/PI cell viability kit (Logos Biosystems, Gyunggi, South Korea) and Luna-FLTM (Logos Biosystems) according to the manufacturer’s protocol. The subpopulations of MNCs were analyzed by flow cytometry, as described above. The blood MNCs and lymph node MNCs stimulated with mitogen, i.e., concanavalin A (ConA; Wako Pure Chemical Industries, Osaka, Japan), or M. bovis (PG45 and five wild strains) were seeded on 96-well tissue culture plates at a concentration of 2 × 105 live MNCs/well. The bovine MNCs were incubated at 37°C in 5% CO2 in the presence of 5 μg of mitogen/well or live M. bovis at a multiplicity of infection (MOI) of 100 for 72 h in triplicate. The cellular proliferative response was measured using a cell counting kit-8 (CCK-8) assay (Dojindo, Osaka, Japan), according to the manufacturer’s protocol.
RNA extraction.
Total RNA (tRNA) extracted from blood MNCs and milk MNCs was obtained using the PureLink RNA mini kit (Ambion, TX, USA). DNase digestion was performed using Turbo DNA-free DNase (Ambion). tRNA was quantified via spectrophotometry using a BioSpec-nano instrument (Shimadzu, Kyoto, Japan). cDNA was synthesized from 1 μg of tRNA using ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) and oligo(dt)18 Primer (Toyobo). For each reaction, a parallel negative-control reaction was performed in the absence of reverse transcriptase and analyzed by the β-actin band using PCR and 1.5% agarose gel stained with ethidium bromide, which was visualized on a UV transilluminator.
Microarray experiment and analysis.
Data from six microarrays (three from day 0 and three from day 7) of blood MNCs following intramammary infusion of M. bovis were provided by TaKaRa Bio Inc. (Shiga, Japan). The gene expression data set was obtained on the Agilent one-color microarray platform (4x44K bovine gene expression array, grid ID 023647). The samples were processed for Agilent microarrays, and data were normalized as previously described (34). A t test was used to identify significant gene expression differences (P < 0.025) between samples. In a further filtering step, only genes with a fold change of ≥2 were selected. bioDBnet was used for gene annotation (35), and GO enrichment analysis was performed using the BioMart enrichment tool (36).
qRT-PCR analysis.
Reverse transcription-quantitative PCR (qRT-PCR) was performed using Thunderbird SYBR qPCR mix (Toyobo) and a MyiQ-icycler (Bio-Rad Laboratories, Hercules, CA, USA). Information on the primers is shown in Table S4 in the supplemental material. Melting curve analysis was used to evaluate each primer pair for specificity to ascertain that only one product was amplified. A BLAST (NCBI platform) search was performed to confirm that the primer sequences amplified only the target gene of interest. Thermal cycling consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The melting temperature of the PCR product was determined by melting curve analysis, which was performed by heating the PCR product from 55°C to 95°C and monitoring the fluorescence change every 0.5°C. The fold-change ratio between preinfection and postinfection samples for each gene was calculated by the ΔΔCT method which is based on the following housekeeping genes: β-actin, tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (37, 38).
Statistical analysis.
Data from the three cows are expressed as the mean ± standard error (SE). Kruskal-Wallis test was used for comparison between groups, Steel test was used for multiple comparisons, and Welch’s t test was used for paired groups, performed using Ekuseru-Toukei 2010 for Windows (Social Survey Research Information, Tokyo, Japan). In all cases, a P value of <0.05 was considered to indicate a statistically significant difference.
Data availability.
The complete microarray data set is publicly available from the ArrayExpress database (accession number E-MTAB-7905).
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev 62:1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maunsell FP, Woolums AR, Francoz D, Rosenbusch RF, Step DL, Wilson DJ, Janzen ED. 2011. Mycoplasma bovis infections in cattle. J Vet Intern Med 25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 3.Nicholas RAJ. 2011. Bovine mycoplasmosis: silent and deadly. Vet Rec 168:459–462. doi: 10.1136/vr.d2468. [DOI] [PubMed] [Google Scholar]
- 4.Fox LK. 2012. Mycoplasma mastitis: causes, transmission, and control. Vet Clin North Am Food Anim Pract 28:225–237. doi: 10.1016/j.cvfa.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Pfützner H, Sachse K. 1996. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders in cattle. Rev Sci Tech 15:1477–1494. doi: 10.20506/rst.15.4.987. [DOI] [PubMed] [Google Scholar]
- 6.Hertl JA, Schukken YH, Welcome FL, Tauer LW, Gröhn YT. 2014. Pathogen-specific effects on milk yield in repeated clinical mastitis episodes in Holstein dairy cows. J Dairy Sci 97:1465–1480. doi: 10.3168/jds.2013-7266. [DOI] [PubMed] [Google Scholar]
- 7.Bannerman DD, Paape MJ, Lee J-W, Zhao X, Hope JC, Rainard P. 2004. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin Diagn Lab Immunol 11:463–472. doi: 10.1128/CDLI.11.3.463-472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannerman DD, Chockalingam A, Paape MJ, Hope JC. 2005. The bovine innate immune response during experimentally-induced Pseudomonas aeruginosa mastitis. Vet Immunol Immunopathol 107:201–215. doi: 10.1016/j.vetimm.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Kauf ACW, Rosenbusch RF, Paape MJ, Bannerman DD. 2007. Innate immune response to intramammary Mycoplasma bovis infection. J Dairy Sci 90:3336–3348. doi: 10.3168/jds.2007-0058. [DOI] [PubMed] [Google Scholar]
- 10.Byrne W, Markey B, McCormack R, Egan J, Ball H, Sachse K. 2005. Persistence of Mycoplasma bovis infection in the mammary glands of lactating cows inoculated experimentally. Vet Rec 156:767–771. doi: 10.1136/vr.156.24.767. [DOI] [PubMed] [Google Scholar]
- 11.Paape M, Mehrzad J, Zhao X, Detilleux J, Burvenich C. 2002. Defense of the bovine mammary gland by polymorphonuclear neutrophil leukocytes. J Mammary Gland Biol Neoplasia 7:109–121. doi: 10.1023/a:1020343717817. [DOI] [PubMed] [Google Scholar]
- 12.Guzman E, Hope J, Taylor G, Smith AL, Cubillos-Zapata C, Charleston B. 2014. Bovine γδ T cells are a major regulatory T cell subset. J Immunol 193:208–222. doi: 10.4049/jimmunol.1303398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gondaira S, Higuchi H, Nishi K, Iwano H, Nagahata H. 2017. Mycoplasma bovis escapes bovine neutrophil extracellular traps. Vet Microbiol 199:68–73. doi: 10.1016/j.vetmic.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 14.Thomas CB, Mettler J, Sharp P, Jensen-Kostenbader J, Schultz RD. 1990. Mycoplasma bovis suppression of bovine lymphocyte response to phytohemagglutinin. Vet Immunol Immunopathol 26:143–155. doi: 10.1016/0165-2427(90)90063-x. [DOI] [PubMed] [Google Scholar]
- 15.van der Merwe J, Prysliak T, Perez-Casal J. 2010. Invasion of bovine peripheral blood mononuclear cells and erythrocytes by Mycoplasma bovis. Infect Immun 78:4570–4578. doi: 10.1128/IAI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanden Bush TJ, Rosenbusch RF. 2003. Characterization of the immune response to Mycoplasma bovis lung infection. Vet Immunol Immunopathol 94:23–33. doi: 10.1016/s0165-2427(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 17.Konnai S, Murata S, Ohashi K. 2017. Immune exhaustion during chronic infections in cattle. J Vet Med Sci 79:1–5. doi: 10.1292/jvms.16-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 19.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 20.Appelberg R. 1992. Mycobacterial infection primes T cells and macrophages for enhanced recruitment of neutrophils. J Leukoc Biol 51:472–477. doi: 10.1002/jlb.51.5.472. [DOI] [PubMed] [Google Scholar]
- 21.Shimabukuro-Vornhagen A, García-Márquez M, Fischer RN, Iltgen-Breburda J, Fiedler A, Wennhold K, Rappl G, Abken H, Lehmann C, Herling M, Wolf D, Fätkenheuer G, Rubbert-Roth A, Hallek M, Theurich S, von Bergwelt-Baildon M. 2017. Antigen-presenting human B cells are expanded in inflammatory conditions. J Leukoc Biol 101:577–587. doi: 10.1189/jlb.5A0416-182R. [DOI] [PubMed] [Google Scholar]
- 22.Jimbo S, Griebel PJ, Townsend H, Babiuk LA, Mutwiri G. 2016. Evidence for the existence of regulatory and effector B cell populations in Peyer’s patches of sheep. Vet Immunol Immunopathol 174:26–34. doi: 10.1016/j.vetimm.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Estes DM. 1996. Differentiation of B cells in the bovine. Role of cytokines in immunoglobulin isotype expression. Vet Immunol Immunopathol 54:61–67. doi: 10.1016/s0165-2427(96)05684-x. [DOI] [PubMed] [Google Scholar]
- 24.Riollet C, Rainard P, Poutrel B. 2000. Kinetics of cells and cytokines during immune-mediated inflammation in the mammary gland of cows systemically immunized with Staphylococcus aureus alpha-toxin. Inflamm Res 49:486–496. doi: 10.1007/s000110050621. [DOI] [PubMed] [Google Scholar]
- 25.Blagitz MG, Souza FN, Batista CF, Diniz SA, Azevedo LF, Silva MX, Haddad JP, Heinemann MB, Cerqueira MM, Della Libera AM. 2015. Flow cytometric analysis: interdependence of healthy and infected udder quarters. J Dairy Sci 98:2401–2408. doi: 10.3168/jds.2014-8727. [DOI] [PubMed] [Google Scholar]
- 26.Mehrzad J, Janssen D, Duchateau L, Burvenich C. 2008. Increase in Escherichia coli inoculum dose accelerates CD8+ T-cell trafficking in the primiparous bovine mammary gland. J Dairy Sci 91:193–201. doi: 10.3168/jds.2007-0096. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas RAJ, Ayling RD. 2003. Mycoplasma bovis: disease, diagnosis, and control. Res Vet Sci 74:105–112. doi: 10.1016/s0034-5288(02)00155-8. [DOI] [PubMed] [Google Scholar]
- 28.Garred P, Honoré C, Ma YJ, Munthe-Fog L, Hummelshøj T. 2009. MBL2, FCN1, and FCN3—the genes behind the initiation of the lectin pathway of complement. Mol Immunol 46:2737–2744. doi: 10.1016/j.molimm.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Ma M, Ippolito GC, Schroeder HW, Carroll MC, Volanakis JE. 2001. Complement activation in factor D-deficient mice. Proc Natl Acad Sci U S A 98:14577–14582. doi: 10.1073/pnas.261428398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belnoue E, Pihlgren M, McGaha TL, Tougne C, Rochat A-F, Bossen C, Schneider P, Huard B, Lambert PH, Siegrist C-A. 2008. APRIL is critical for plasmablast survival in the bone marrow and poorly expressed by early-life bone marrow stromal cells. Blood 111:2755–2764. doi: 10.1182/blood-2007-09-110858. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi H, Iwano H, Gondaira S, Kawai K, Nagahata H. 2011. Prevalence of Mycoplasma species in bulk tank milk in Japan. Vet Rec 169:442. doi: 10.1136/vr.d5331. [DOI] [PubMed] [Google Scholar]
- 32.Gondaira S, Higuchi H, Iwano H, Nakajima K, Kawai K, Hashiguchi S, Konnai S, Nagahata H. 2015. Cytokine mRNA profiling and the proliferative response of bovine peripheral blood mononuclear cells to Mycoplasma bovis. Vet Immunol Immunopathol 165:45–53. doi: 10.1016/j.vetimm.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Kiku Y, Ozawa T, Kushibiki S, Sudo M, Kitazaki K, Abe N, Hideyuki T, Hayashi T. 2010. Decrease in bovine CD14 positive cells in colostrum is associated with the incidence of mastitis after calving. Vet Res Commun 34:197–203. doi: 10.1007/s11259-009-9339-8. [DOI] [PubMed] [Google Scholar]
- 34.Chain B, Bowen H, Hammond J, Posch W, Rasaiyaah J, Tsang J, Noursadeghi M. 2010. Error, reproducibility and sensitivity: a pipeline for data processing of Agilent oligonucleotide expression arrays. BMC Bioinformatics 11:344. doi: 10.1186/1471-2105-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudunuri U, Che A, Yi M, Stephens RM. 2009. bioDBnet: the biological database network. Bioinformatics 25:555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, Bardou P, Beck T, Blake A, Bonierbale M, Brookes AJ, Bucci G, Buetti I, Burge S, Cabau C, Carlson JW, Chelala C, Chrysostomou C, Cittaro D, Collin O, Cordova R, Cutts RJ, Dassi E, Di Genova A, Djari A, Esposito A, Estrella H, Eyras E, Fernandez-Banet J, Forbes S, Free RC, Fujisawa T, Gadaleta E, Garcia-Manteiga JM, Goodstein D, Gray K, Guerra-Assunção JA, Haggarty B, Han D-J, Han BW, Harris T, Harshbarger J, Hastings RK, Hayes RD, Hoede C, et al. . 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res 43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson TL, Sutherland IA, Sutherland J. 2007. Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol 115:160–165. doi: 10.1016/j.vetimm.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Spalenza V, Girolami F, Bevilacqua C, Riondato F, Rasero R, Nebbia C, Sacchi P, Martin P. 2011. Identification of internal control genes for quantitative expression analysis by real-time PCR in bovine peripheral lymphocytes. Vet J 189:278–283. doi: 10.1016/j.tvjl.2010.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete microarray data set is publicly available from the ArrayExpress database (accession number E-MTAB-7905).