Figure 6. Model.
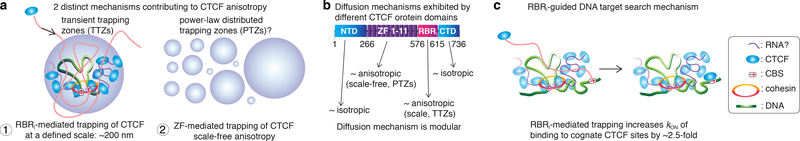
(a) Two distinct mechanisms contribute to CTCF anisotropy. First, RBRi-mediated trapping of CTCF in transiently trapping zones (TTZs) of a characteristic size (~200 nm). TTZs are likely to correspond to clusters of CTCF that form in a largely RBRi-dependent manner37 and therefore, perhaps, are held together by RNA(s) of a currently unknown identity. Cognate Binding Sites (CBSs) reside within the TTZ, as does a piece of DNA strand (green). Second, ΔRBRi-CTCF still displays scale-free anisotropy, which may be due to non-specific interactions with DNA. We speculatively model this as arising from trapping in power-law distributed zones (PTZs).
(b) Diffusion mechanism is modular. By mutating individual protein domains, the effect of each individual domain can be determined. CTCF contains 4 major protein domains: an N-terminal domain (NTD) of largely unknown function, 11 Zinc Fingers essential for DNA-binding, a short internal RNA-binding region (RBRi), and a C-terminal domain (CTD) of largely unknown function. The ZF-domain and the RBRi-domain appear to mediate anisotropic diffusion via PTZ and TTZ type mechanisms, respectively.
(c) Target-search mechanism. The RBRi increases the apparent rate at which CTCF locates a cognate DNA-binding site. Given that the RBRi also mediates CTCF clustering, we speculate that RBRi-mediated CTCF clusters, perhaps near loop boundaries, help “guide” diffusing CTCF towards its cognate DNA binding sites (CBSs; DNA is shown in green).