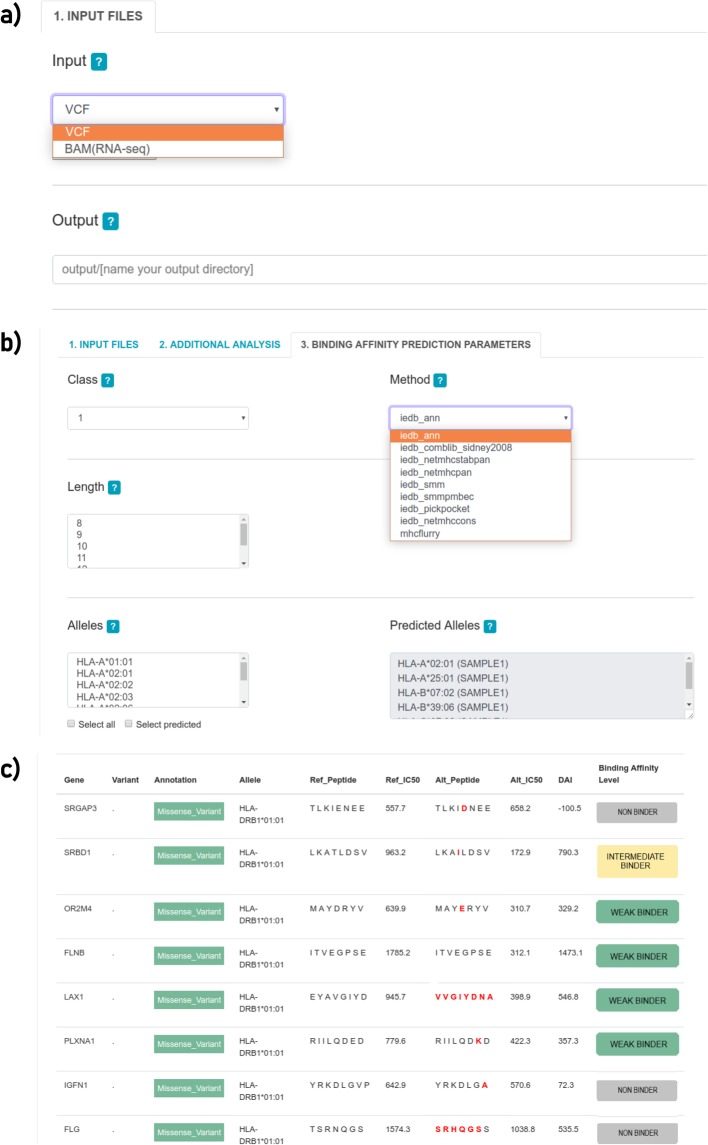
Fig. 2.
Screenshots of neoANT-HILL interface. a Processing tab for submitting genome or transcriptome data. b Processing tab for parameters selection to run neoepitope binding affinity prediction. On this tab, all the parameters can be defined by the users through selection boxes, ranging from the MHC class, corresponding prediction methods, to parallelization settings. The length and HLA alleles parameters allow multiple selections, although that might interfere in the processing time. c Binding prediction results tab shows an interactive table which reports all predicted neoepitopes and information about each prediction, respectively. The interactive table shows several columns, such as the donor gene, HLA allele, mutation type, reference (Ref_Peptide) and altered (Alt_peptide) peptides sequences, reference (Ref_IC50) and altered (Alt_IC50) binding affinity scores, binding affinity category (High, Moderate, Low, and Non-binding) and differential agretopicity index (DAI)