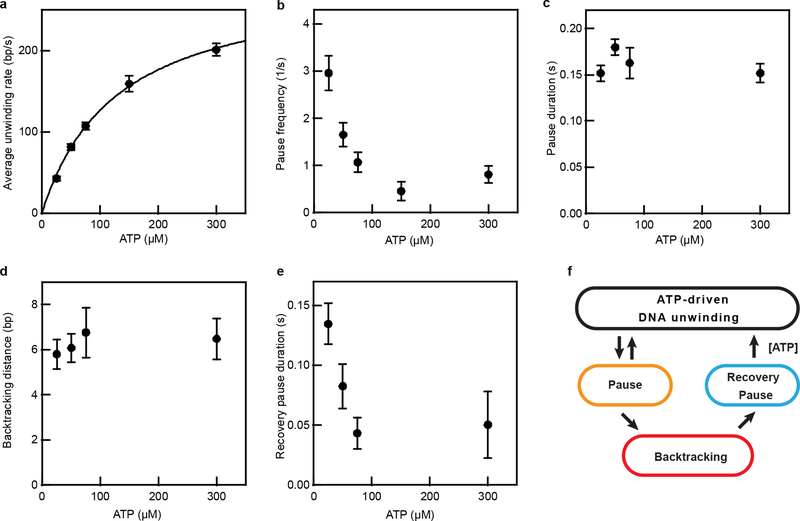
Extended Data Figure 4 |. RecBCD unwinding rate and pausing characteristics at solution pH 6.
a, Average unwinding rate as a function of ATP concentrations fit to a Michaelis-Menten dependence with vmax = 290 ± 10 bp/s, KM = 130 ± 10 μM. Data are mean ± s.e.m (n = 47, 94, 80, 37, 110 trajectories from at least three independent biological replicates for each condition). b, ATP concentration dependence of the pause frequency. Pause frequency was determined as the average number of pauses per second for each single-molecule trajectory. Data are mean ± s.e.m. (n = 47, 94, 80, 37, 110 trajectories, from at least three independent biological replicates for each condition). c, Median duration of pauses during forward unwinding at various ATP concentrations. The error bars are the s.d. of the median derived from resampling (n = 76, 62, 27, 18 events, from at least three independent biological replicates for each condition). d, Mean backtracking distance at various ATP concentrations. Data are mean ± s.e.m (n = 20, 22, 16, 10 events, from at least three independent biological replicates for each condition). e, Median recovery pause duration after a backtracking event at various ATP concentrations. The error bars are the s.d. of the median derived from resampling. The p-values for the differences between the 25 μM ATP data point and the 50, 75, and 300 μM ATP data points are 0.05, 0.004, and 0.09, respectively, derived from two-sided Kolmogorov-Smirnov test of the distributions of the pause durations (n = 20, 22, 16, and 10 events for the 25, 50, 75, and 300 μM points, respectively). Note that not all trajectories contain a backtracking event. Data acquired at 10% glycerol, solution pH 6, 500 Hz. f, Schematic of a kinetic model of RecBCD-induced DNA unwinding. During DNA unwinding, pausing occurs frequently and some pauses lead to enzyme backtracking; the enzyme can exit the backtracking state and resume DNA unwinding through a recovery pause intermediate, which is distinct from the initial pause state.