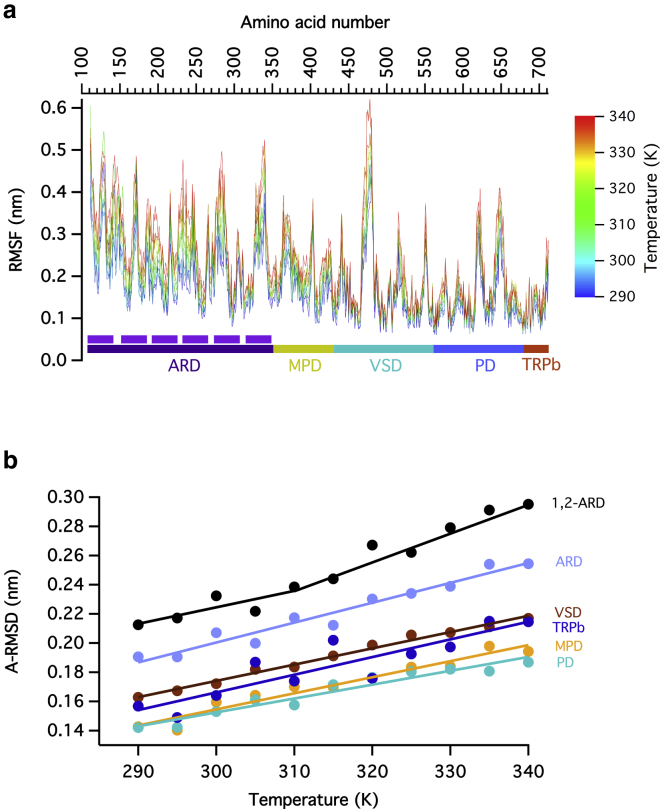
Figure 2.
Analysis of the structural fluctuations induced by temperature. (a) RMSF of the beads that correspond to the main chain of each amino acid in coarse-grained molecular dynamics, calculated over different subunits and different replicas for each simulated temperature. The ANKs are indicated by purple bars. The ANKs that have the greatest RMSF are ANK1 and ANK2. (b) The calculated average value of the RMSD (A-RMSD) of each functional domain during the last 500 ns of each simulated replica and each of the four subunits at a given temperature is shown as a function of the simulation temperature. The data is fit to a linear function. Notice that the ARD 1–2 data has an increased slope at 310 K. To see this figure in color, go online.