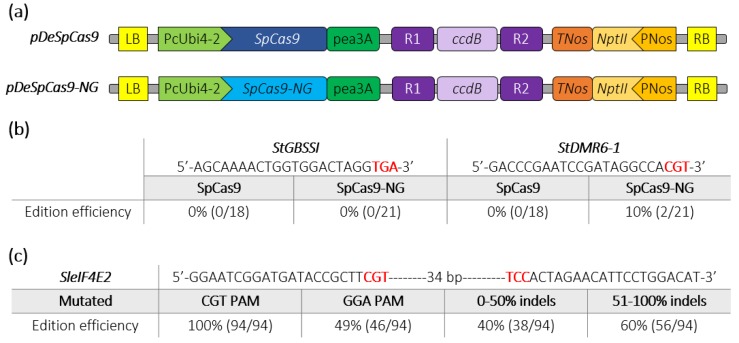
Figure 2.
Schematic representation of SpCas9 and SpCas9-NG binary plasmids and editing efficiencies in potato and tomato. (a) The construct backbones used for Agrobacterium-mediated transformation of potato (Desiree) and tomato (WVA106) are schematically represented. The R1_ccdB_R2 cassette allows the insertion of the guide cassette through a Gateway LR reaction. (b) Edition efficiency represents the percentage of mutated potato plants among the transgenic plants. The PAM sequence is depicted in bold red. (c) Among the mutated tomato plants screened by HRM, 94 plants were Sanger sequenced and analyzed using ICE analysis. Edition efficiency represents the percentage of mutated plants with the SpCas9-NG at the SleIF4E2 locus for each target (CGT or GGA PAMs). The mutated plants are classified into two groups according to the rate of indels at the targeted eIF4E2 locus. The PAM sequence is depicted in bold red.