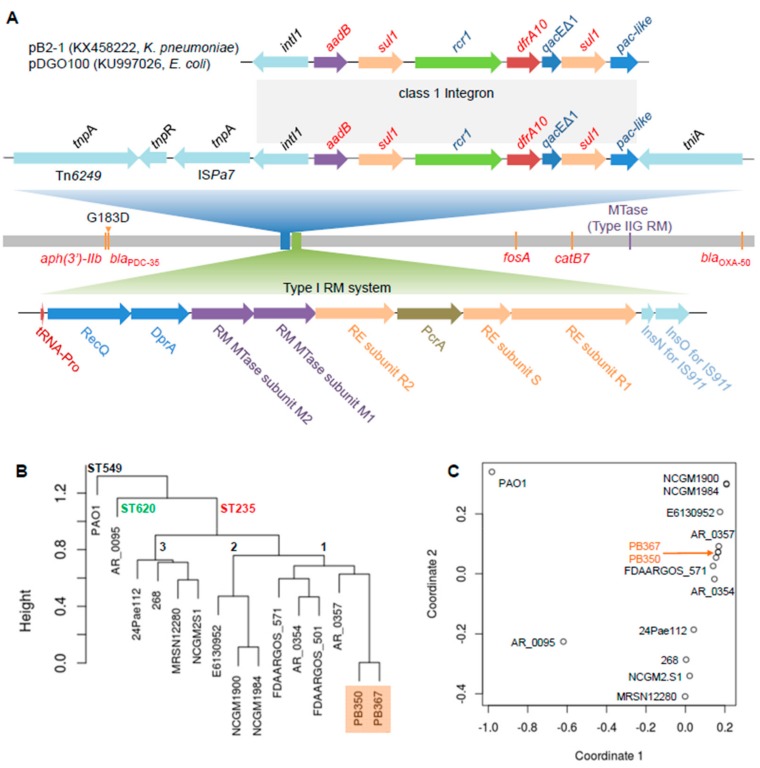
Figure 1.
Comparative genome analysis of P. aeruginosa isolates. (A) Schematic genome of the paired P. aeruginosa isolates, PB367 and PB350. Genomic mutation C931,543T (G183D) in β-lactamase gene (blaPDC-35) from PB367 to PB350 is indicated in the chromosome, along with other antimicrobial resistance genes (aph(3’)-IIb, fosA, catB7, and blaOXA-50) and a gene encoding a putative Type IIG restriction-modification (RM) methyltransferase (MTase) targeting a specific motif AAGm6AYC. Top: A unique class 1 integron in P. aeruginosa with multidrug resistance (MDR) genes, 100% identical to the ones found in plasmids of K. pneumoniae and E. coli, is located among multiple mobile genetic elements, Tn6249, resolvase, ISPa7, and tniA. Bottom: A unique Type I RM system comprised of restriction enzymes (RE, subunits R and S) and modification MTases (M1 and M2) putatively targeting a bipartite and asymmetric motif CTm6AC(N)5GGG/Cm4CC(N)5GTAG; (B) Phylogeny tree of 15 P. aeruginosa isolates, mainly ST235, from kWIP hierarchical clustering using the complete linkage method. PAO1 belonged to ST549, AR_0095 belonged to ST620, and three main sub-ST235 clusters (1–3) are demonstrated; (C) Relatedness overview of 15 P. aeruginosa isolates in a plot of metric multidimensional scaling (MDS).