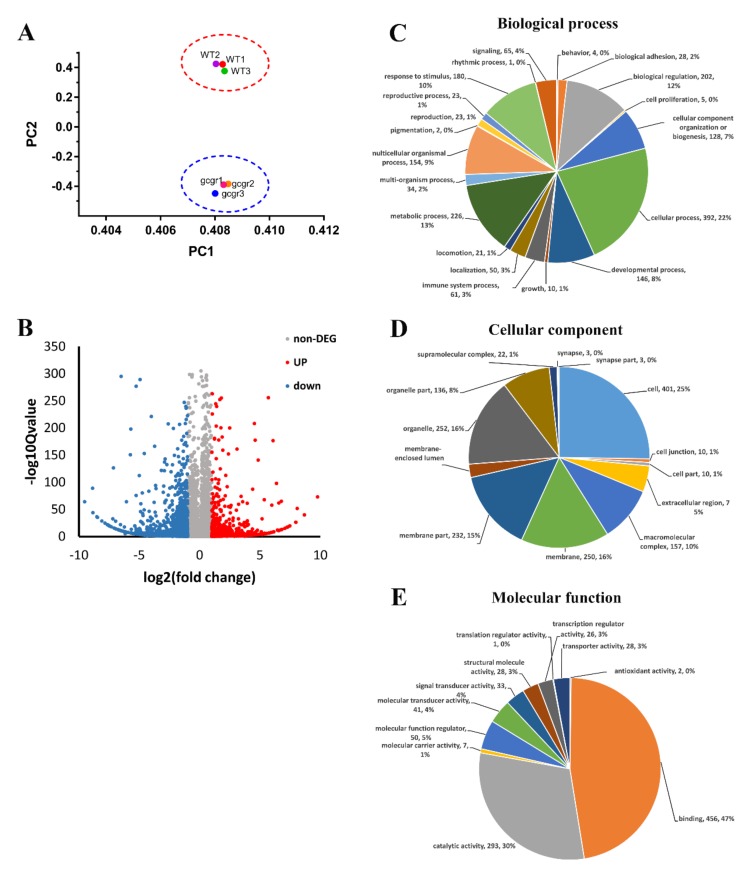
Figure 1.
RNA-seq (RNA sequencing) analysis of glucagon receptor (gcgr−/−) mutant zebrafish. (A) Principal component analysis (PCA) plot of three wildtype and gcgr−/− mutant RNA-seq datasets. Principal component 1 (PC1), and principal component 2 (PC2) were used for analysis. (B) Volcano plot of differential expression analysis of gcgr−/− mutant and control larvae showing the relationship between p-value and log fold changes. Red shows upregulated genes and blue downregulated genes. (C–E) Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs) in gcgr−/− mutant zebrafish. The DEGs were assigned to three categories: Biological process, (C) cellular component, (D) and molecular function (E). The names of the GO subcategories, the number of genes, and the proportion of each subcategory are listed by the pie charts.