Figure 1.
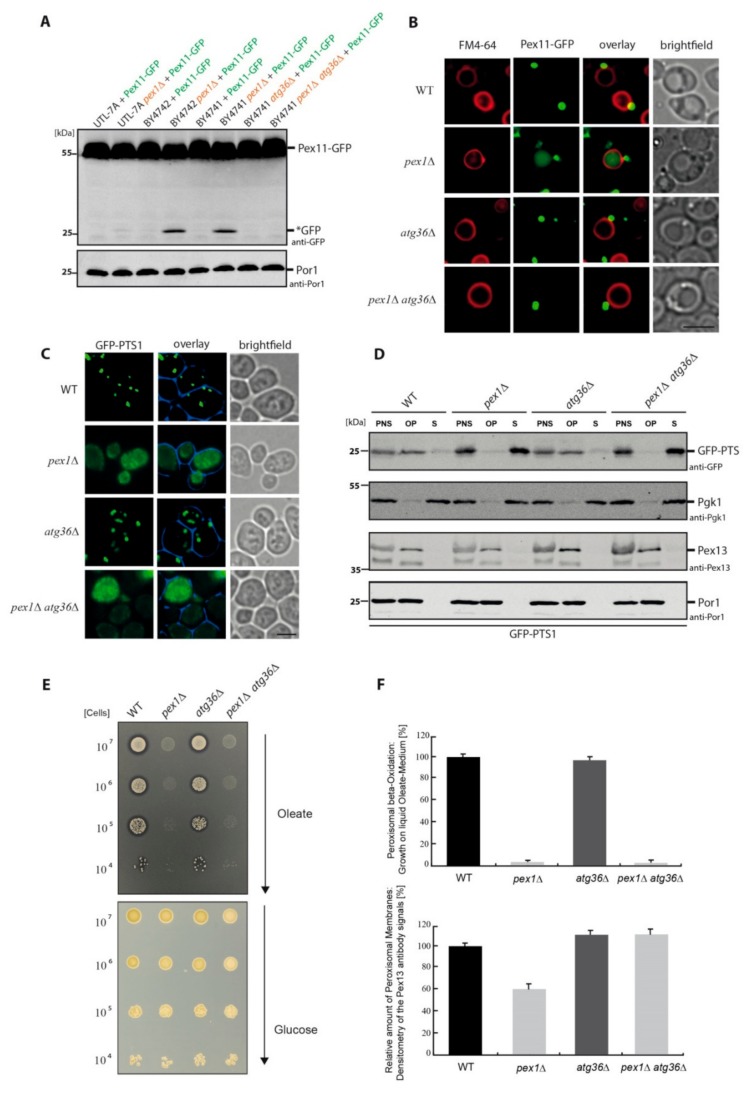
The matrix protein import defect in PEX1-deficient cells occurs independently of pexophagy. (A) The indicated different Saccharomyces cerevisiae wild-type (WT) strains were transformed with the peroxisomal membrane protein Pex11 genetically fused to green fluorescent protein (GFP). The autophagic degradation of peroxisomes is shown by the occurrence of free *GFP. The pex1Δ strains of the BY4741 and BY4742 background display a constitutive degradation of peroxisomes, while the UTL-7A pex1Δ strain lacks PEX1 deletion-induced pexophagy. The degradation of peroxisomes depended on the presence of Atg36. The mitochondrial protein Por1 was used as loading control. Uncropped versions of the blots can be seen in the Supplemental Figure S1A. (B) The vacuolar membrane of BY4741 cells was stained red with FM4-64, while peroxisomal structures labeled with Pex11-GFP are visible as green dots. In addition, the pex1Δ strain displays a diffuse green staining within the vacuole, demonstrating that a portion of the peroxisome population was degraded via pexophagy. This degradation is fully blocked in the pex1Δatg36Δ double mutant. Bar: 5µm. (C) Cells were transformed with a plasmid encoding the peroxisomal matrix protein marker GFP-PTS1. Cells with punctate pattern displayed a functional import, while cytosolic mislocalization indicated an import defect. Bar: 5µm. (D) The subcellular sedimentation analysis of the prepared post-nuclear supernatant (PNS) showed that the matrix protein GFP-PTS1 can mainly be detected in the organellar pellet (OP) fraction of WT and atg36Δ cells (intact import), while it is mislocalized to the cytosolic supernatant (S) fraction in pex1Δ and pex1Δatg36Δ cells (import defect). The level of the endogenous peroxisomal membrane protein Pex13 was elevated in cells without Atg36. The mitochondrial Por1 and the cytosolic Pgk1 served as controls. Uncropped versions of the blots can be seen in the Supplemental Figure S1B. (E) The indicated strains were spotted as a series of 10-fold dilutions on a glucose medium as well as on a medium with oleate as the sole carbon source. The WT and atg36Δ strains had an intact peroxisome biogenesis and could grow on oleate plates. The utilization of oleate during beta-oxidation was further indicated by the formation of halos around the drop spots. The pex1Δ and pex1Δatg36Δ strains were both unable to grow on oleate medium, indicating a defect in beta-oxidation and peroxisome function. (F) Lower diagram: The densitometry data from the Pex13-positive antibody signals from Western blots of organellar pellet fractions were compared. The block of pexophagy in pex1Δatg36Δ resulted in a rise of the Pex13-level of the pex1Δ strain back to WT level. Upper diagram: The functionality of the indicated strains in beta-oxidation was analyzed by monitoring cell growth in liquid oleate medium (n = 3). The value for PEX1-deficient cells did not improve with the additional deletion of ATG36, suggesting that the block of pexophagy does not improve peroxisomal function in beta-oxidation.