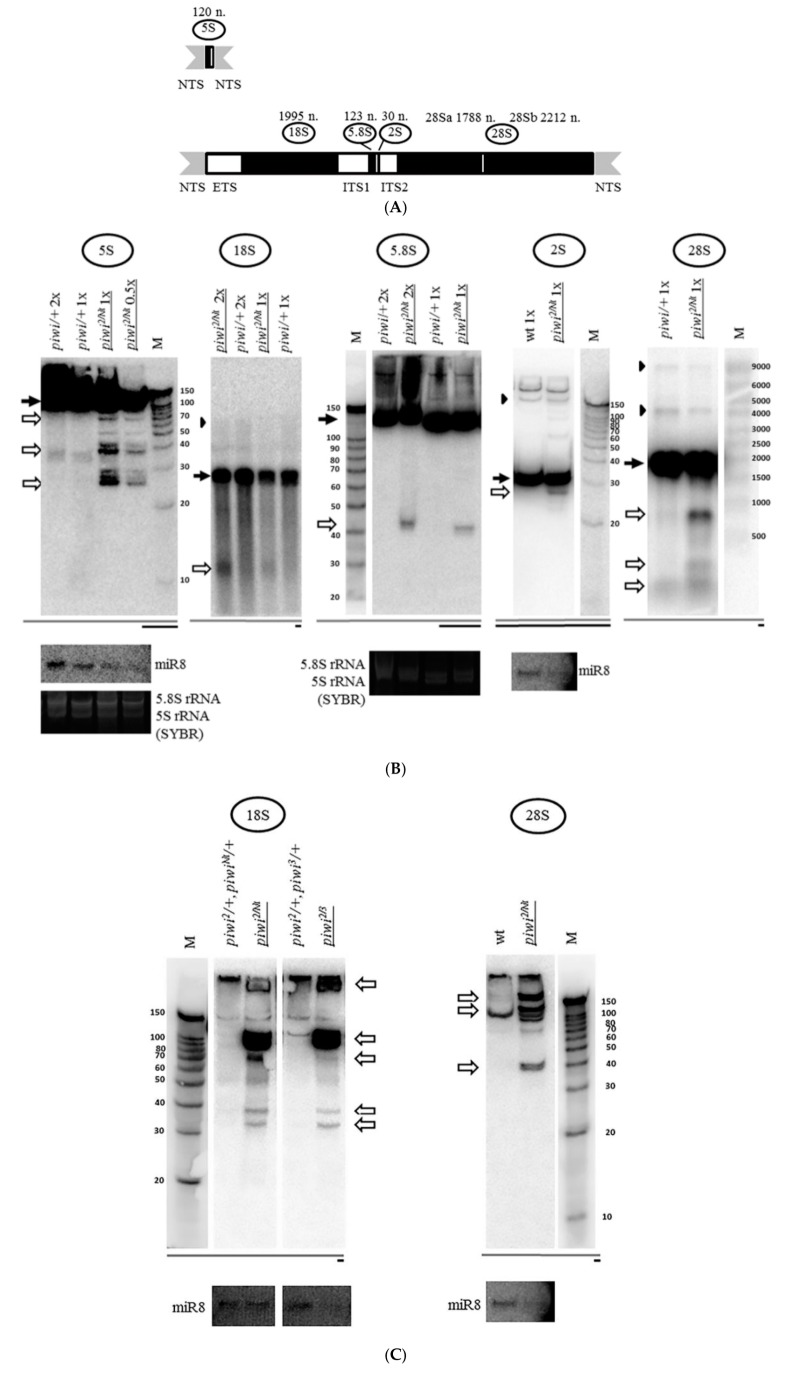
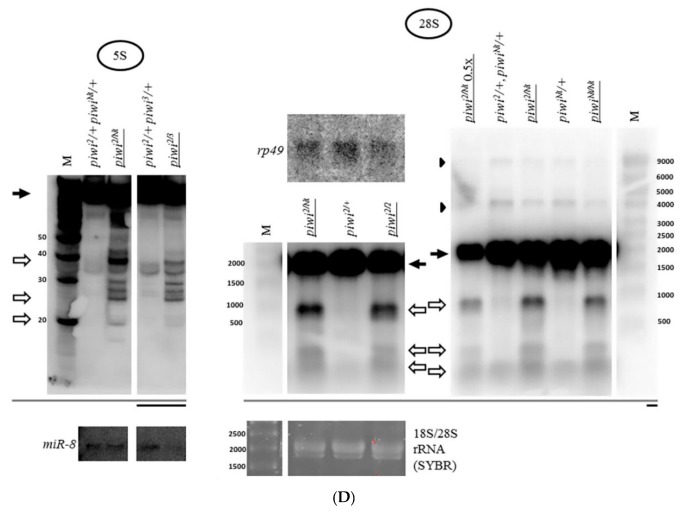
Figure 1.
Mutation in the piwi gene causes the accumulation of fragments of all rRNAs in Drosophila melanogaster ovaries: (A) Schematic representation of a repeat of the tandem 5S rDNA cluster (top) and a repeat of the tandem 18S/28S rDNA cluster (bottom) in Drosophila melanogaster. The only ribosomal RNA polymerase III transcript, 5S rRNA, is produced as a precursor of 135 n. from the tandem 5S rDNA repeats. Nucleolar rRNAs are transcribed by RNA polymerase I as a single pre-rRNA precursor 8093 n. in size from the tandem 18S/28S rDNA repeats, processed to produce 18S, 5.8S, 2S, and 28S rRNA (divided into two parts—28Sa and 28Sb). Regions encoding mature rRNAs are shown in black and their sizes are indicated above. Transcribed spacers are shown in white: ETS: external transcribed spacer; ITS1, and ITS2: internal transcribed spacers 1 and 2. Nontranscribed spacers are shown in gray: NTS: nontranscribed intergenic spacer. (B) Transheterozygous combination piwi2/Nt leads to the accumulation of fragments of all rRNAs in Drosophila melanogaster ovaries. Shown are the results of Northern blot analysis of ovarian RNA with probes henceforth selected to detect the 3′-terminus of an rRNA species (except Figure 2 and Supplementary Figure S3). In the case of 2S rRNA the whole rRNA is detected, as its length is equal to the length of the probe (30 n.). The positions of the probes are henceforth depicted in coarse schemes below the Northern blot images as black bars with mature species shown as dark grey bars. The effect of accumulation of fragments (henceforth white arrows) is observed for all rRNAs. In most cases, the signals of mature rRNAs (henceforth black arrows) are brought to relative saturation, so that the fragments can be seen more clearly. rRNA precursors are marked with black arrowheads where apparent. The analyzed genotypes are indicated above the lanes (with mutant genotypes henceforth underlined): piwi2/Nt and control piwi/+, where piwi/+ is henceforth a mixture of heterozygous siblings piwi2/+ and piwiNt/+. The wt genotype did not contain a mutation in piwi (nos-GAL4 driver line). Equal amounts of total RNA were loaded per lanes to be compared, which is indicated by the symbols 2×, 1×, or 0.5× for relative RNA quantity. Rehybridization with a probe to detect miR-8 microRNA or total RNA staining by SYBR Green II of the corresponding gels before blotting (labeled as SYBR) was used to control for loading. M henceforth indicates the lane with a marker, marker sizes in nucleotides are indicated. (C) For long 18S and 28S rRNAs, in addition to large fragments, smaller fragments are also detected in piwi mutants. For 18S (left) and 28S rRNA (right), fragments of the size range from short RNAs and above are observed. (D) The fact of rRNA fragment accumulation does not depend on a specific defect in the piwi gene. Fragments accumulate not only in transheterozygous piwi2/Nt and homozygous piwiNt/Nt ovaries where Piwi has lost its nuclear localization (exemplified by 28S rRNA on the right), but also in the strong transheterozygous piwi2/3 combination (exemplified by 5S rRNA on the left) and in null homozygous piwi2/2 ovaries (exemplified by 28S rRNA in the middle). To all the lanes of each blot equal amounts of total RNA was loaded or a 0.5× amount where indicated. Rehybridization with probes to detect miR-8 microRNA and ribosomal protein rp49 mRNA or total RNA staining by SYBR Green II of the corresponding gels before blotting henceforth was used to control for loading. Drosophila long rRNAs have similar sizes of 1778 n. for 28Sa, 1995 n. for 18S, and 2212 n. for 28Sb, which frequently yields two signals in 1% agarose gels upon SYBR Green II staining.