Figure 6.
Identifying a Core Set of TRα1 Target Genes in GABAergic Neurons of the Striatum by Combining RNAseq and Chip-Seq Analyses
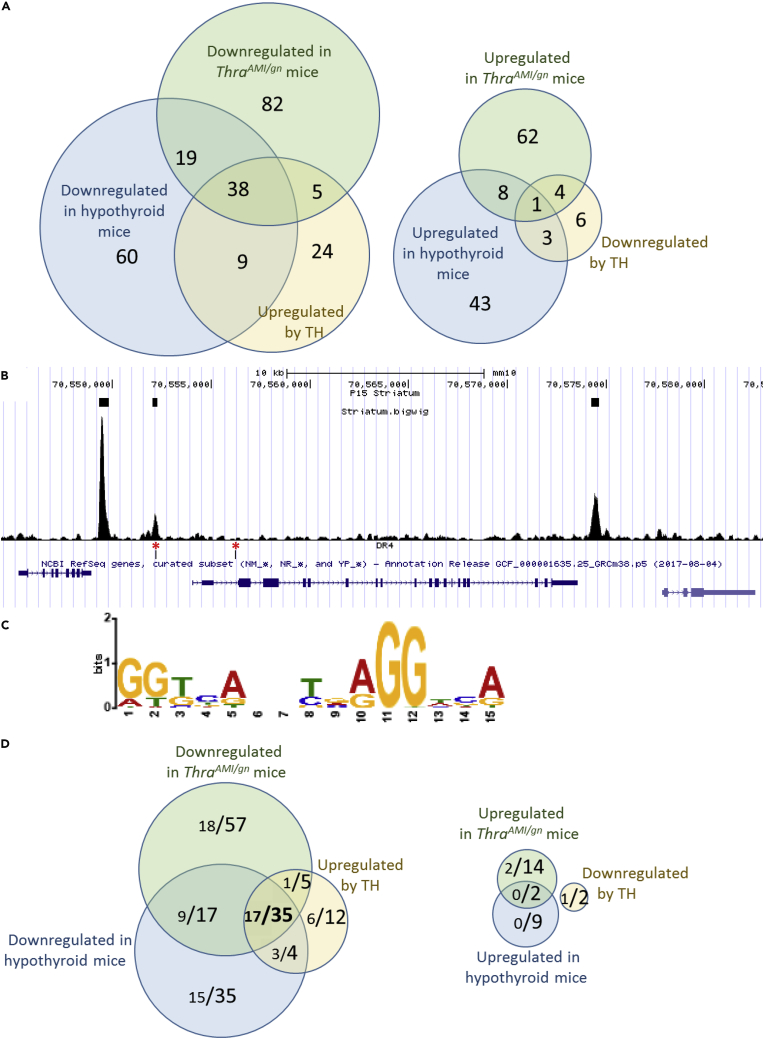
(A) RNAseq identifies a set of 38 genes, the expression pattern of which is fully consistent with a positive regulation by TRα1, and only 1 gene that has the opposite expression pattern.
(B) Extract of the Mus musculus genome browser, around the Hr gene, a well-characterized TRα1 target gene. The 3 upper boxes indicate TRBSs identified as significant by the MACS2 algorithm. Note that a DR4-like element (lower track, red asterisks), as defined below, is found in only one of the 3 peaks.
(C) Consensus sequence found in TRBSs identified by de novo motif search is close to the DR4 consensus (5′AGGTCANNNNAGGTCA-3′).
(D) Combinations of RNAseq and Chip-Seq data. In the Venn diagrams, each fraction gives the number of genes with a proximal TRBS (<30 kb for transcription start site, large lettering) and, among these genes, those in which a DR4 element was identified (small lettering). A set of 35 genes fulfill the criteria for being considered as genuine TRα1 target genes: they are downregulated in hypothyroid and mutant mice and upregulated after TH treatment of hypothyroid mice. For 17 of these genes, the TRBS contains a recognizable DR4-like element.
See also Data S1 and Tables S2–S5.