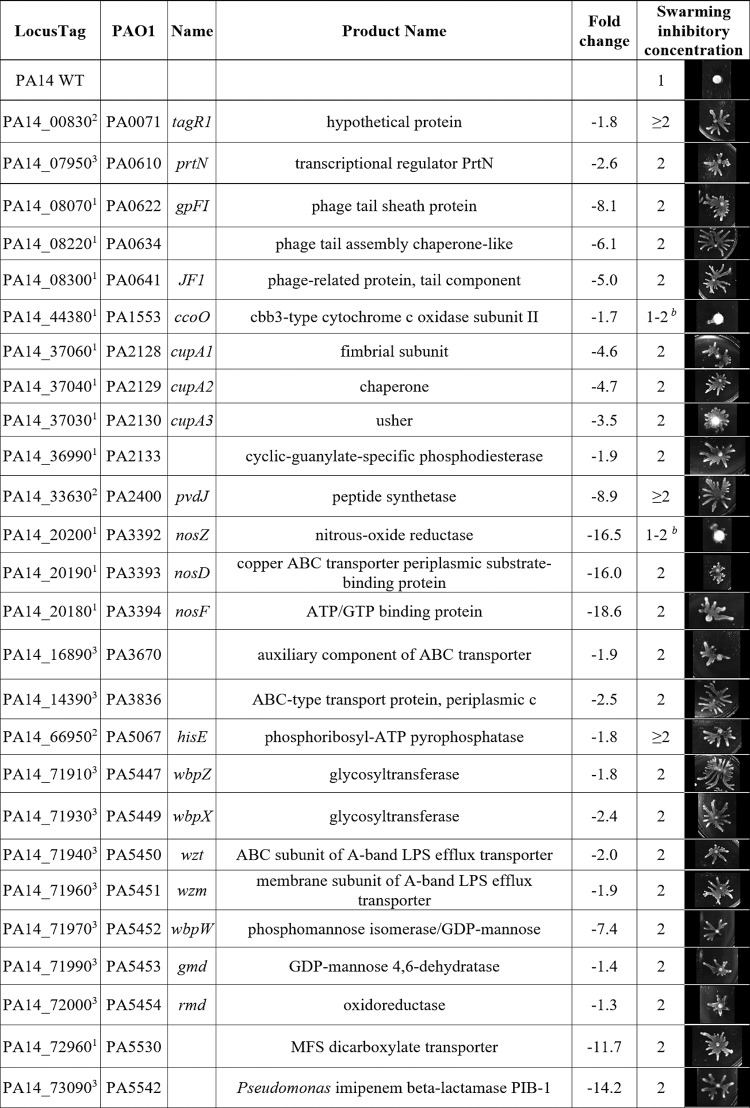
TABLE 2.
Genes dysregulated under swarming conditions that matched the known resistome revealed 26 tobramycin resistance mutantsa
See references 8–13. PA14 transposon mutants in selected genes were tested for altered tobramycin susceptibility under swarming conditions using the agar dilution method (inhibitory concentrations shown in micrograms per milliliter tobramycin) along with images of swarming colonies at 1 μg/ml. Evidence of dysregulation came from swarm versus swim RNA-Seq (1) or tobramycin RNA-Seq (2). Selected genes were also confirmed by qRT-PCR from reference 15 (3). Additional mutants in genes showing no evidence of dysregulation (gmd and rmd) but belonging to operons containing dysregulated genes were also tested. Seventeen additional mutants are described in Table S3 in the supplemental material.
Two mutants showed minimal swarming at tobramycin 1 μg/ml but grew better than WT at 2 μg/ml.