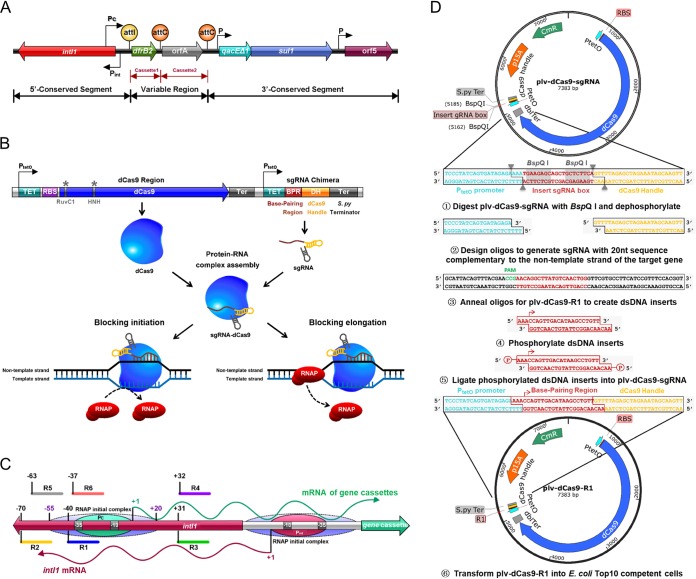
FIG 1.
Schematic diagram of engineering a CRISPRi system to repress a class 1 integron in E. coli. (A) Structural diagram of a class 1 integron on conjugative plasmid R388. (B) Harnessing CRISPRi to block transcription initiation and elongation. RNAP, RNA polymerase. (C) Genetic organization of intI1, gene cassettes, Pc promoter, Pint promoter, and the binding sites of six sgRNAs. The six sgRNAs were designed for targeting different regions around the Pc promoter, and all target sites were located in the IntI1 integrase-coding region. The transcription start site is labeled as +1. Arabic numerals highlight the distance to the transcription start site of gene cassettes. The dotted purple oval shows the initial RNA polymerase complex. The green oval and red oval represent the Pc promoter of gene cassettes and the Pint promoter of intI1, respectively. (D) Protocol for construction of CRISPRi recombinant plasmids. The same strategy is adopted to target the different regions of R388 class 1 integron in E. coli. The nucleotides shown with a gray background are BspQI recognition sequences, and gray triangles indicate the BspQI cutting sites. Red arrowheads indicate the transcription start sites of the base-pairing region of sgRNA.