Fig. 3.
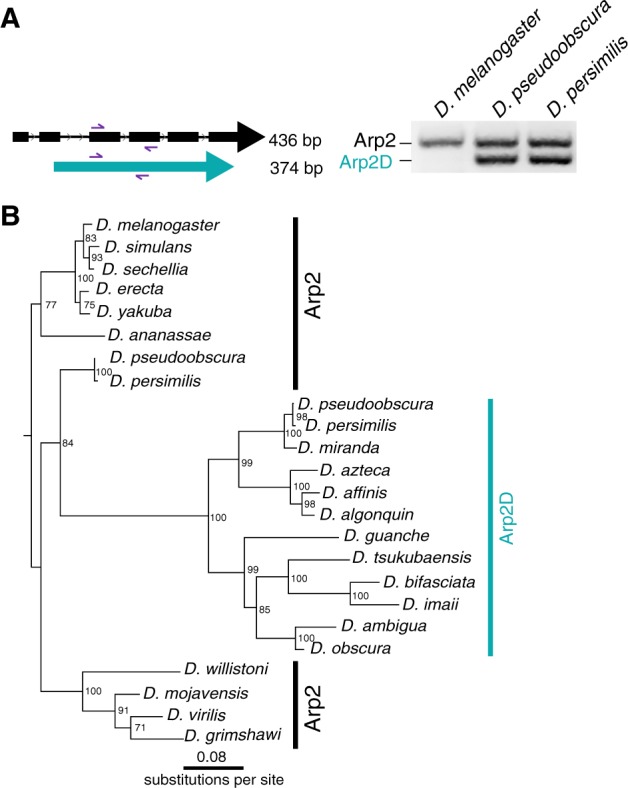
Arp2D originated from a retroduplication event of Arp2. (A) Primers targeting sequences in two neighboring exons conserved in Drosophila pseudoobscura Arp2 and Arp2D were used to PCR-amplify the genomic region in D. melanogaster, D. pseudoobscura, and D. persimilis. PCR products were run on a 2% agarose gel. As Arp2D does not contain introns, the PCR product is smaller (374 bp) than that of Arp2 (436 bp). (B) Nucleotide sequences from Arp2 (from 12 sequenced Drosophila species) and Arp2D sequences from the obscura group were aligned (no gaps were removed). A PhyML tree (Guindon et al. 2010) with 100× resampling was generated and presented using the species from the D. melanogaster subgroup as an outgroup.
