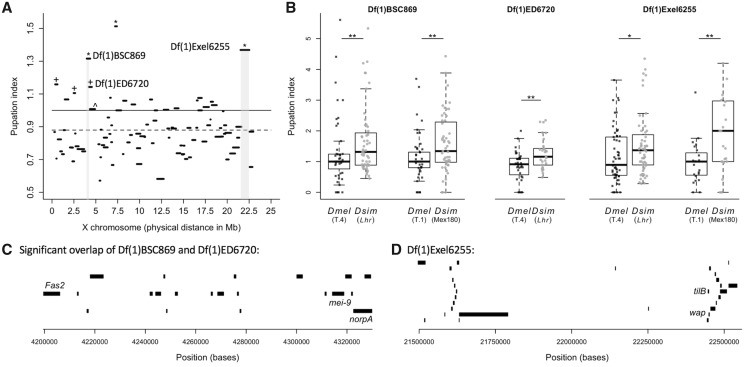
Fig. 4.
Hybrid deficiency screen of the X chromosome identifies two regions of interest. (A) The median pupation index for each of the 90 deficiency hybrid crosses is plotted by the physical map distance each engineered deletion spans along the X chromosome. Deficiencies with a pupation index significantly >1 (solid line) after correction for multiple comparisons are denoted by an asterisk (sample sizes and significance levels for all deficiencies are listed in supplementary table S3A, Supplementary Material online). Deficiencies with a median pupation index significantly >0.88 (the grand mean, dashed line) after correction for multiple comparisons are denoted with a cross. All of these lines also had pupation indices significantly >1 before correcting for multiple comparisons. The two regions we pursued for candidate gene validation are highlighted in light gray. Note that we did not pursue the remaining three significant deficiency strains (from left to right: +Df(1)BSC530, +DF(1)ED411, *Df(1)6906) because they showed similar pupation indices when crossed to Drosophila melanogaster (supplementary fig. S4, Supplementary Material online). (B) The pupation indices of the deficiencies from the gray highlighted areas in (A) are shown for the original hybrid cross (Lhr) and for a cross to the T.4 D. melanogaster strain. BSC869 and Exel6255 were additionally crossed to the D. simulans strain Mex180 and the D. melanogaster strain T.1. Asterisks denote a significant difference between pupation indices of deficiency strains crossed to D. melanogaster and D. simulans (Wilcoxon tests; *P < 0.05, **P < 0.01). Sample sizes can be found in supplementary table S3A, Supplementary Material online. (C) The region uncovered by the overlap of deficiencies Df(1)BSC869 and Df(1)ED6720, but excluding the region uncovered by Df(1)ED6727 (which had a pupation index no different than 1; marked in (A)with ^) and the 23 genes contained within it. The three genes with available disruption strains (Fas2, mei-9, and norpA) are labeled. (D) The region uncovered by Exel6255 and the 28 genes contained within it. The two genes with available disruption strains (tilB and wap) are labeled.