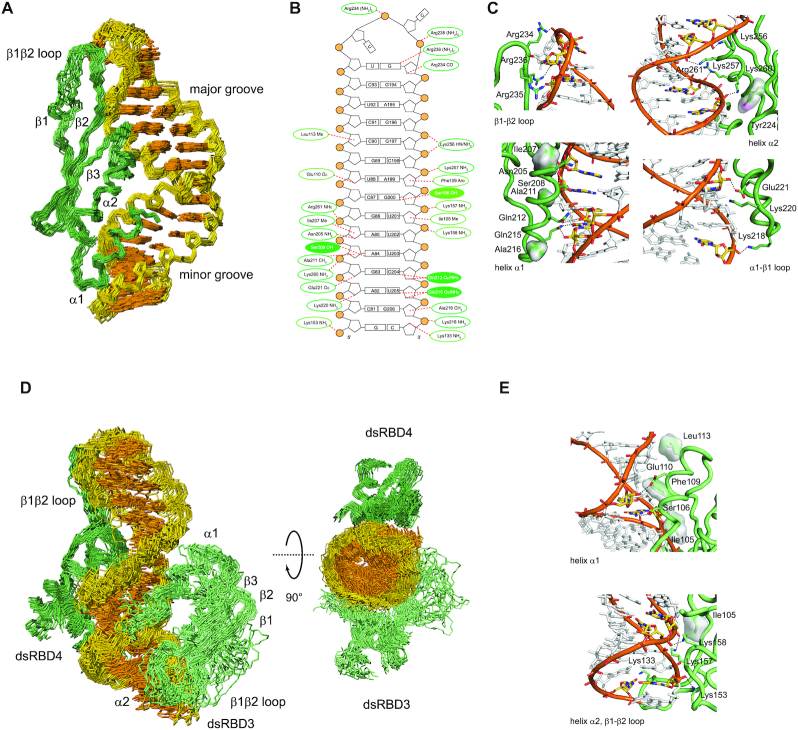
Figure 2.
Structure of STAU1 dsRBD4 and dsRBD3/4 in complex with sARF1 SBS dsRNA. (A) Structural ensemble of the STAU1 dsRBD4–sARF1 SBS dsRNA complex. Heavy-atom superposition of the ensemble of the 20 lowest-energy structures (Table 2). The protein backbone is shown in dark green and the RNA heavy atoms of the bases in orange and those of the ribose-phosphodiester backbone are shown in gold (omitting phosphate and 2′-OH oxygens). (B) Schematic representation of sARF1 SBS dsRNA showing interactions of dsRBD3 as well as dsRBD4 as dotted lines. Interactions with the ribose-phosphodiester backbone are circled in dark green for dsRBD4 and light green for dsRBD3 while base interactions are shown as filled circles. (C) Details of the interactions of dsRBD4 β1–β2 loop, helix α2, helix α1 and α1–β2 loop with sARF1 SBS dsRNA. Amino acid side chains mediating interactions with the dsRNA are shown as sticks, interacting dsRNA residues are shown in yellow with nitrogen atoms in blue and oxygen atoms in red and the phosphodiester backbone is in orange with the phosphate oxygens in red. Protein residues making hydrophobic contacts are shown with their Van der Waals surface and hydrogen bonds are indicated by black dotted lines. (D) Structural ensemble of the STAU1 dsRBD3/4–sARF1 SBS dsRNA complex. Heavy-atom superposition of the ensemble of the 20 lowest-energy structures (Table 2). The protein backbone of dsRBD4 is shown in dark green, the one of dsRBD3 in light green and the RNA heavy atoms of the bases in orange and those of the ribose-phosphodiester backbone are shown in gold (omitting phosphate and 2′-OH oxygens). A second view rotated by 90° along the horizontal axis shows that dsRBD3 and dsRBD4 interact with sARF1 SBS dsRNA on opposite sides of the helix. The linker residues (172–204) are omitted. (E) Details of the interactions of dsRBD3 helix α1, helix α2 and β1–β2 loop with sARF1 SBS dsRNA. Color scheme as in C.